The H7N9 virus underwent dynamic evolution during infection within individual patients. Signature amino acids in PB2 and NA demonstrated high polymorphism, and signature sites related to antiviral resistance and mammalian adaption underwent dynamic variation during disease progression.
Keywords: H7N9, dynamic variation, R292K, E627K
Abstract
Background
Signature amino acids of H7N9 influenza A virus play critical roles in human adaption and pathogenesis, but their dynamic variation is unknown during disease development.
Methods
We sequentially collected respiratory samples from H7N9 patients at different timepoints and applied next-generation sequencing (NGS) to the whole genome of the H7N9 virus to investigate the variation at signature sites.
Results
A total of 11 patients were involved, from whom 29 samples were successfully sequenced, including samples from multiple timepoints in 9 patients. Neuraminidase (NA) R292K, basic polymerase 2 (PB2) E627K, and D701N were the 3 most dynamic mutations. The oseltamivir resistance-related NA R292K mutation was present in 9 samples from 5 patients, including 1 sample obtained before antiviral therapy. In all patients with the NA 292K mutation, the oseltamivir-sensitive 292R genotype persisted and was not eliminated by antiviral treatment. The PB2 E627K substitution was present in 18 samples from 8 patients, among which 12 samples demonstrated a mixture of E/K and the 627K frequency exhibited dynamic variation. Dual D701N and E627K mutations emerged but failed to achieve predominance in any of the samples.
Conclusions
Signature amino acids in PB2 and NA demonstrated high polymorphism and dynamic variation within individual patients during H7N9 virus infection.
Since its first report in spring 2013, the novel avian H7N9 virus has continued to circulate in China and has caused 5 waves of human infection [1]. Unlike the highly pathogenic avian influenza (HPAI) H5N1 virus, the original avian H7N9 is weakly pathogenic to poultry and spreads silently in poultry populations, increasing the chances for human exposure and leading to human infections [2]. The novel H7N9 virus is undergoing rapid evolution, and several HPAI H7N9 strains were recently identified in human cases and poultry outbreaks [3, 4]. As of 30 October 2017, there have been >1565 laboratory-confirmed human cases of avian influenza H7N9 infection in China, with a fatality rate of approximately 39% [5].
The H7N9 virus shows distinct biological characteristics compared to the HPAI H5N1 virus [6]. H7N9 hemagglutinin (HA) demonstrates a dual receptor profile, binding to both α2,3- and α2.6-linked sialic acid, which allows it to replicate efficiently in both the upper and lower human respiratory tracts [7]. H7N9 virus shows moderate transmissibility among both ferrets and guinea pigs in transmission evaluation studies [8]. Accumulating evidences have indicated that signature amino acids in the HA and basic polymerase 2 (PB2) play an important role for the avian H7N9 virus to cross species barrier and for mammalian adaption [7–10]. Moreover, some amino acid substitutions in the neuraminidase (NA) segment significantly reduce the inhibitory activity of oseltamivir in infected ferrets [11]. These molecular markers are critical for viral pathogenesis and related to clinical outcome of human infection [12, 13].
Due to the low fidelity of its RNA polymerase, the replication of influenza virus is error prone and the virus population in a host is not a genetically homologous colony, but a collection of viruses coupled by diverse mutations [14]. This community of closely coupled viruses is termed a viral quasispecies. Quasispecies analysis could detect dynamic variation in the virus signature amino acids during disease development. Notably, the oseltamivir-resistant NA R292K mutation increased during antiviral treatment in a fatal case of H7N9 infection [12], and the PB2 E627K mutation was also identified with increasing frequency during infection in a fatal human case of avian H7N7 influenza virus [15]. Thus, these molecular markers undergo dynamic evolution during human infection. However, the kinetics of their variation throughout the whole infection stage is unknown at present. In this study, we sequentially collected respiratory samples throughout disease progression from patients infected with avian H7N9 influenza virus and performed deep sequencing of the virus genomes amplified directly from clinical samples. Our results described the dynamic variation in signature amino acids across the whole genome of the H7N9 virus during human infection.
METHODS
Patients and Samples
We included 11 patients diagnosed with A/H7N9 infection admitted to the China–Japan Friendship Hospital (4 cases), the First Affiliated Hospital of Soochow University (4 cases), and the First Affiliated Hospital of Nanchang University (3 cases). Table 1 outlines the demographic details, comorbidities, antiviral and corticosteroid treatments received, and final disposition of each patient. All patients had received antiviral treatment with oseltamivir after disease onset, and 5 patients also received peramivir therapy. After admission and confirmation of infection, clinical samples were sequentially collected per day from patients during disease development, till the day there was no virus shedding in the respiratory tract, tested by real-time reverse-transcription polymerase chain reaction (RT-PCR). The study protocol was approved by the Medical Ethical Committee of all 3 hospitals, and written informed consent was obtained from all subjects who participated in the study and/or their families.
Table 1.
Demographic Details, Therapy, and Outcome of Patients With A(H7N9) Infection Involved in This Study
| Case No. | Hospital | Sex | Age, y | Comorbidities | Date of Disease Onset | Time From Disease Onset to Admission, d | Date of Antiviral Therapy | Antiviral Regimen | Corticosteroid (Methylprednisolone) (Time After Disease Onset) | Outcome (Time After Disease Onset) |
|---|---|---|---|---|---|---|---|---|---|---|
| Mild group | ||||||||||
| 1 | CJFH | M | 48 | None | 31 Jan 2017 | 8 | 21 Feb 2017 | Oseltamivir 150 mg bid | None | Discharge (day 31) |
| 2 | FANU | M | 67 | Diabetes | 1 Feb 2017 | 7 | 6 Feb 2017 | Oseltamivir 150 mg bid; peramivir 300 mg qd | 40 mg bid (day 7–13) | Discharge (day 27) |
| 3a | FASU | F | 66 | Diabetes; RA; osteoporosis | 9 Jan 2017 | 5 | 14 Jan 2017 | Oseltamivir 150 mg bid; peramivir 600 mg bid | 40 mg qd (day 5–10) | Discharge (day 27) |
| Severe group | ||||||||||
| 4a | FASU | F | 59 | Postoperation of thymoma | 16 Dec 2016 | 8 | 24 Dec 2016 | Oseltamivir 150 mg bid | 40 mg bid (day 8–14) | Death (day 16) |
| 5ab | FANU | M | 81 | Hypertension; diabetes | 28 Jan 2017 | 11 | 6 Feb 2017 | Oseltamivir 150 mg bid; peramivir 600 mg bid | 40 mg bid (day 8–12) | Death (day 15) |
| 6a | FASU | F | 61 | COPD | 18 Dec 2016 | 7 | 27 Dec 2016 | Oseltamivir 150 mg bid | 40 mg qd (day 7–17) | Death (day 19) |
| 7a | FANU | F | 69 | Atrial fibrillation | 27 Jan 2017 | 9 | 4 Feb 2017 | Oseltamivir 150 mg bid; peramivir 600 mg bid | 40 mg bid (day 20–25) | Death (day 28) |
| 8a | FASU | M | 67 | Silicosis; hypertension; cerebral infarction | 7 Jan 2017 | 4 | 11 Jan 2017 | Oseltamivir 150 mg bid; peramivir 300 mg qd | 40 mg qd (day 4–10) | Death (day 13) |
| 9ab | CJFH | M | 16 | None | 2 June 2017 | 1 | 6 June 2017 | Oseltamivir 150 mg bid | 320–200 mg qd (day 26–29)c, 160–40 mg qd (day 30–40) | Discharge (day 135) |
| 10ab | CJFH | M | 31 | None | 10 June 2017 | 6 | 16 June 2017 | Oseltamivir 150 mg bid; peramivir 600 mg bid | 320–200 mg qd (day 19–22)c, 80–40 mg qd (day 22–27) | Discharge (day 109) |
| 11a | CJFH | F | 55 | None | 2 Apr 2017 | 15 | 19 Apr 2017 | Oseltamivir 150 mg bid | 80 mg qd (day 25–27) | Death (day 44) |
Abbreviations: bid, twice daily; CJFH, China–Japan Friendship Hospital; COPD, chronic obstructive pulmonary disease; F, female; FAUN, First Affiliated Hospital of Nanchang University; FASU, First Affiliated Hospital of Soochow University; M, male; qd, once daily; RA, rheumatoid arthritis.
aMechanical ventilation was applied to this patient.
bExtracorporeal membrane oxygenation was applied to this patient.
cOrganizing pneumonia developed in cases 9 and 10, and high doses of corticosteroids were used first and lowered as clinical condition relieved later.
Next-Generation Sequencing
Viral RNA was extracted from respiratory samples using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), and the whole genome was amplified with 3 universal primers targeting the conserved termini of influenza A virus, as previously described [16, 17]. The PCR products were sheared, end-repaired, A-tailed, and ligated to Illumina sequencing adaptors containing molecular identifier tags using the NEBNext DNA Library Prep reagent set (NEB, Ipswich, Massachusetts) to allow multiplex sequencing per lane. The libraries were pooled, clustered, and sequenced on an Illumina HiSeq 2500 platform (TruSeq version 3 chemistry) with a paired-end 500-cycle sequencing protocol with indexing. Ten samples achieved insufficient reads, and their PCR products were resequenced with the same protocol in a second run.
Bioinformatics Analysis
Next-generation sequencing (NGS) data analysis was conducted with CLC Genomic Workbench 10.1.1 software (Qiagen). First, a trimming step was performed to remove reads with an error rate >1% or reads with a length <200 bp. As signal calling in the terminal base was error-prone, we trimmed 20 bases at the 5ʹ end and 1 at the 3ʹ end for all reads. The reads passing quality control were assembled de novo with the “de novo assembly” tool in the CLC package under the parameters “word size = 20, bubble size = 50.” After assembly and scaffolding, contigs with lengths >800 bp were extracted and blasted against an H7N9 genome sequence collection downloaded from the National Center for Biotechnology Information (NCBI) Influenza Virus Database (accessed 1 October 2017). The 8 segments with the highest scores in the blast hits were selected as references for reads mapping using the CLC mapping tool with parameters set as match score = 1, mismatch score = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.8, and similarity fraction = 0.8. After mapping, a consensus sequence was extracted from the mapping result for each segment with every base covered by at least 10 reads (depth >10×). When disagreement happened on the base at a given position, the vote option was selected to make sure the majority of the reads decide which base was recalled. The consensus sequences of HA, NA, and PB2 of the avian H7N9 virus were used for phylogenetic analysis. Sequences available from NCBI and GISAID (Global Initiative on Sharing Avian Influenza Data) were downloaded to construct neighbor-joining trees with the Kimura 2-Parameter model and 1000 bootstraps using Mega 7 software [18].
The basic variant detection tool in the CLC package was used for variant detection from the mapping result of each segment with defaulted parameters but adjusted the minimum coverage = 10, minimum count = 3, minimum frequency = 1% (calculated as “count/coverage”). The variant nucleotides were then transformed into amino acid variants using standard genetic code. We used the χ2 test to compare amino acid variations among different timepoints with SPSS 20.0 (IBM SPSS, Chicago, Illinois). Detailed methods for bioinformatics analysis are provided in the Supplementary Appendix.
Validation Test of the NA R292K Mutation
We used a single-nucleotide polymorphism (SNP)–specific RT-PCR assay to validate the NA R292K mutation detected by NGS [19]. The reaction was performed by using a SensiFAST probe No-ROX One-Step Kit (Bioline, London, United Kingdom) with 4 µL of RNA added as template. The assay was performed on a LightCycler 480 Instrument II, and the data were processed by using LightCycler 480 SW 1.5.1 software (Roche, Basel, Switzerland).
RESULTS
Evolutionary Relationships of H7N9 Virus In Vivo
We included 11 patients aged 16–81 years in our study. All patients developed pneumonia, and 9 patients required mechanical ventilation, among whom 3 patients also required extracorporeal membrane oxygenation (ECMO) (Table 1). Although all patients were treated with NA inhibitors (NAIs), 6 patients ultimately died, including 1 who received ECMO support.
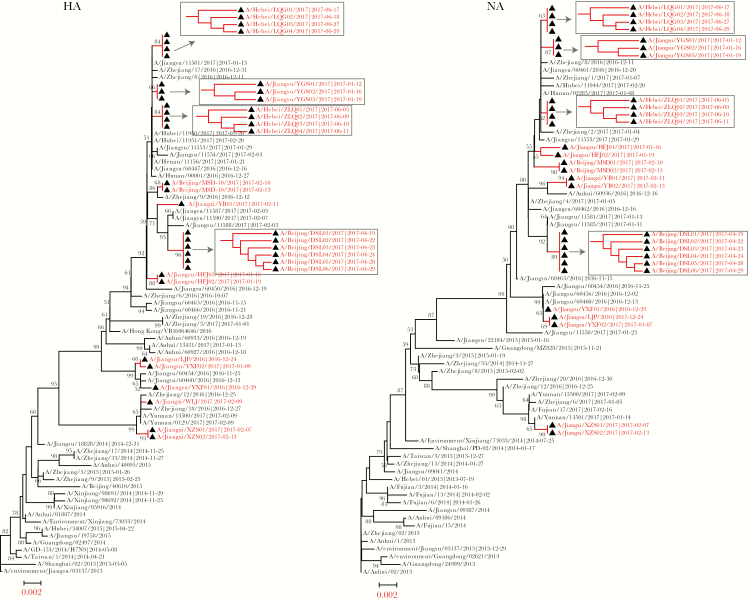
Viral genome information was obtained from a total of 29 samples by NGS, and 9 patients had samples from >1 timepoint (Table 2). The HA and NA clustered with those of viruses isolated in the Yangtze Delta Region from late 2016 to early 2017 (Figure 1). Specifically, HA and NA were grouped with viruses isolated from Jiangsu and Zhejiang provinces, although several cases in our study were from the north of China (cases 1 and 8–10). Moreover, all viral HA and NA showed similar topological structures, in which the virus from a specific patient were located in a similar position in both phylogenetic trees. Viruses from the same person obtained on different dates formed a single cluster and had the same branch length. When topology alone was investigated, viruses from the early timepoint occupied a superior evolutionary position and served as the ancestors of later viruses (Figure 1, cases 8–11).
Table 2.
Signature Amino Acid Variation of Influenza A(H7N9) Viruses During Disease Developmenta
| HA (H3 Numbering) | NA (N2 Numbering) | PB2 | PB1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Favor Mammalian Receptor Binding | NAI Resistance | Increased Virulence in Mammalian Models | Increased Transmission in Ferrets | ||||||||
| Case No. | Sample Date | Virus Name (Sample Type) | T160A | G186V/I | Q226L/I/P | H274Y | R292K | K526R | E627K | D701N | V473L |
| 1 | 10 Feb 2017 | A/Beijing/MSD01/2017(BALF) | A | G | L | H | R | 27 b R | 103E | 3063D/337N | 91V |
| 13 Feb 2017 | A/Beijing/MSD02/2017(BALF) | A | G | L | H | R | R | 173E | 14749D/1017N | 51V | |
| 2 | 11 Feb 2017 | A/Jiangxi/YB01/2017 (NP) | 168A | V | 174 L/46P | H | 183K/324R | R | E | D | V |
| 13 Feb 2017 | A/Jiangxi/YB02/2017 (NP) | 13A | c | c | 122H | 113R | c | c | c | c | |
| 3 | 16 Jan 2017 | A/Jiangsu/HFJ01/2017 (NP) | A | G | L | H | R | R | 959K/43E | D | V |
| 19 Jan 2017 | A/Jiangsu/HFJ02/2017 (NP) | 58A | 71G | 81L | 13H | 12R | 20R | 73K | D | V | |
| 4 | 24 Dec 2016 | A/Jiangsu/LJP/2016 (NP) | 62A | 80V | 81L | 139H | R | 92K | 61K/30E | 4994D/396N | 104V |
| 5 | 9 Feb 2017 | A/Jiangxi/WLJ/2017 (NP) | 1090T/17880A | V | L | H | 15367K/11 192R | K | E | D | V |
| 6 | 29 Dec 2016 | A/Jiangsu/YXF01/2016 (Sputum) | A | V | L | 140H | R | 36K | 21K/40E | 19673D/147N | 70V |
| 7 Jan 2017 | A/Jiangsu/YXF02/2017 (Sputum) | 155A | 198V | L | 60H | 110R | 66K | 50K | 277D/16N | 67V | |
| 7 | 7 Feb 2017 | A/Jiangxi/XZS01/2017 (NP) | 419T/1174A | V | I | 2548H/857Y | 2107K/1446R | K | K | D | 1633V/192I |
| 13 Feb 2017 | A/Jiangxi/XZS02/2017 (NP) | 4T/10A | 22V | 25I | 68H | 68K/33R | c | c | c | c | |
| 8 | 12 Jan 2017 | A/Jiangsu/YGS01/2017 (NP) | A | V | I | H | R | K | E | 68343N/1844D | V |
| 16 Jan 2017 | A/Jiangsu/YGS02/2017 (NP) | A | V | L | H | R | 50K | 172E | 3478N/208D | 86V | |
| 19 Jan 2017 | A/Jiangsu/YGS03/2017 (NP) | A | V | L | H | R | 90K | 52E | 1161N/41D | 131V | |
| 9 | 5 June 2017 | A/Hebei/ZLQ01/2017 (BALF) | A | 3471I/642V | L | H | 8779K/1312R | K | K | D | V |
| 9 June 2017 | A/Hebei/ZLQ02/2017 (BALF) | A | 415I/67V | L | H | 867K/265R | c | c | c | V | |
| 10 June 2017 | A/Hebei/ZLQ03/2017 (BALF) | A | 3536I/746V | L | H | 1800K/693R | K | K | D | V | |
| 11 June 2017 | A/Hebei/ZLQ04/2017 (BALF) | 181A | 204I/28V | L | 121H | 92K/23R | 20K | K | D | 38V | |
| 10 | 17 June 2017 | A/Hebei/LQG01/2017 (BALF) | A | V | L | H | R | 106K | 164K/43E | D | V |
| 18 June 2017 | A/Hebei/LQG02/2017 (BALF) | A | V | L | H | R | 30K | 145K/47E | D | V | |
| 27 June 2017 | A/Hebei/LQG03/2017 (BALF) | A | V | L | H | R | 51K | 123K/35E | D | V | |
| 29 June 2017 | A/Hebei/LQG04/2017 (BALF) | 32A | 36V | 46L | 47H | 66R | c | c | c | c | |
| 11 | 19 Apr 2017 | A/Beijing/DSL01/2017 (sputum) | A | V | L | H | 1,654R | K | 356K/135E | D | 130V |
| 22 Apr 2017 | A/Beijing/DSL02/2017 (sputum) | A | V | L | H | 2393R/66K | K | 289K/402E | D | 145V | |
| 23 Apr 2017 | A/Beijing/DSL03/2017 (sputum) | A | V | L | H | 656R | 171K | 98K/73E | D | 83V | |
| 24 Apr 2017 | A/Beijing/DSL04/2017 (sputum) | A | V | L | H | R | K | 331K/133E | D | 112V | |
| 28 Apr 2017 | A/Beijing/DSL05/2017 (sputum) | 83A | 111V | 125L | H | R | 46K | 78K/45E | D | c | |
| 29 Apr 2017 | A/Beijing/DSL06/2017 (sputum) | 50A | 75V | 92L | H | R | 66K | 35K/70E | D | c | |
Abbreviations: BALF, bronchoalveolar lavage fluid; HA, hemagglutinin; NA, neuraminidase; NAI, neuraminidase inhibitor; NP, nasopharyngeal swab; PB1, basic polymerase 1; PB2, basic polymerase 2.
aSignature amino acid sites listed in the table must be covered by at least 10 reads (depth ≥10×). Variants below this depth were omitted. Signature amino acids prone to mammalian adaption or transmission are labeled with bold font.
bCounts of mapped reads to this amino acid (depth ≤200×, or coexistence of heterologous amino acids). Sites without number attached means its depth was >200×.
cSites were not covered with enough reads (<10 reads) and variant calling was unavailable.
Figure 1.
Phylogenetic analysis of hemagglutinin (HA) and neuraminidase (NA) of influenza A(H7N9) virus from the patients in this study. Bootstrap values are marked in the tree nodes, and values <50% are hidden in the trees. Viruses from this study are labeled with a black triangle and colored red. Viruses from the same patient at different timepoints were further investigated in a topology subtree.
Drug Resistance Mutations in NA During Antiviral Treatment
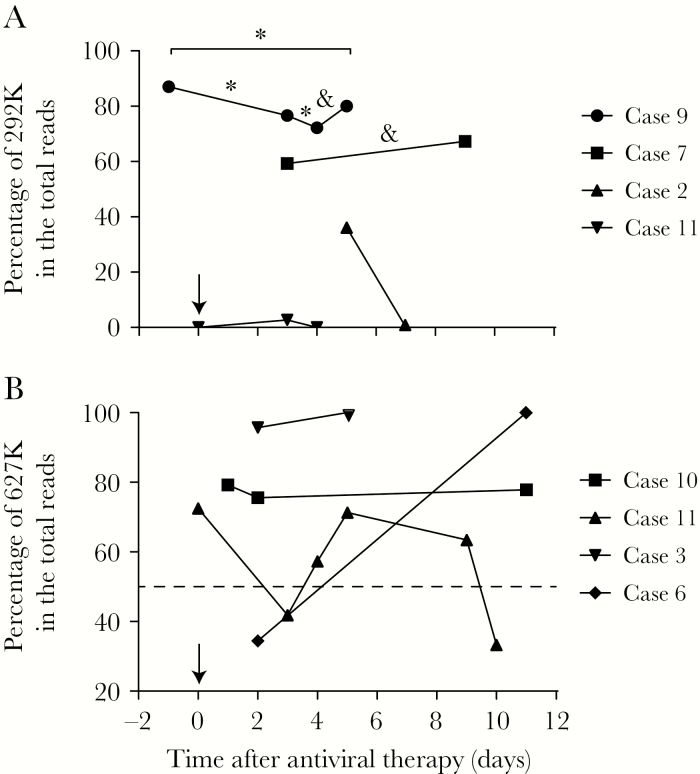
We conducted nucleotide polymorphism analysis of the sites related to NAI resistance in the NA segment, including R118K, E119V, D151E, R152K, I222V, R224K, H274Y, E276D, R292K, N294S, and R371K (N2 numbering). R292K, the most prevalent mutation, was observed in 9 samples from 5 patients (Table 2). In all the samples with 292K identified, wild-type (WT) 292R persisted and was not overwhelmed or eliminated at the final timepoint. In patients who were sampled at several timepoints, the R292K mutation expressed dynamic variation (Figure 2A). In case 2, the R292K mutation was detected in 183 reads (36%) 5 days after NAI treatment but was not detected 2 days later. A similar situation happened with case 11; this patient received antiviral treatment on April 19, 2017, and no mutation was observed at this timepoint. Then, the R292K substitution was detected in 2.7% of 2459 total reads on 22 April but faded by the next day. Cases 7 and 9 each had a mixed 292K/R population that was sustained over 6 days during NAI treatment. In case 7, 2107 reads (59.5%) acquired the R292K mutation 3 days after antiviral therapy, but the proportion of NAI-resistant genotypes only slightly increased to 67.3% with another 6 days of NAI treatment, and there was no significant difference (Figure 2A). In case 9, 87.0% of the reads in the bronchoalveolar lavage fluid obtained on 5 June contained the R292K mutation; at this timepoint, the laboratory test for H7N9 was not complete, and antiviral treatment was not yet implemented. The patient was given NAI antiviral therapy starting on the second day, and the ratio of resistant reads slightly decreased to 76.6% and 72.2% on the third and fourth days, respectively. Throughout 6 days of antiviral treatment, the drug-resistant genotype did not eliminate the WT but expressed a downward trend (Figure 2A). Moreover, we validated the R292K mutation in these samples with a SNP RT-PCR assay [19]. All samples were positive in the assay targeting the H7 segment, but 2 samples, A/Jiangxi/YB02/2017 in case 2 and A/Beijing/DSL02/2017 in case 11, were negative in the assays targeting NA 292K/R and NA292K, respectively (Table 3).
Figure 2.
The kinetics of R292K and E627K mutations in sequential samples obtained from patients during disease development. A, The percentage of 292K reads exhibiting variation in 4 patients at multiple timepoints. A χ2 test was applied to cases 7 and 9 (underlined). *Significant (P < .05 with 95% confidence interval) differences between 2 timepoints. &Insignificant differences between 2 timepoints. B, The percentage of variation in 627K reads in sequential samples collected from 4 cases. The day antiviral therapy was initiated was defined as day “0” and marked with a black arrow. The dashed line indicates the position of 50% on the y-axis.
Table 3.
Reverse-Transcription Polymerase Chain Reaction Validation of the R292K Mutation Detected by Next-Generation Sequencing
| Case No. | Sample Date | Virus Name | No. of Reads | Cycle Threshold Valuea | |||
|---|---|---|---|---|---|---|---|
| 292K | 292R | H7 | 292K | 292R | |||
| 2 | 11 Feb 2017 | A/Jiangxi/YB01/2017 | 183 | 324 | 32.17 | 34.83 | 34.91 |
| 13 Feb 2017 | A/Jiangxi/YB02/2017 | 0 | 113 | 37.36 | Noneb | None | |
| 5 | 9 Feb 2017 | A/Jiangxi/WLJ/2017 | 15367 | 11192 | 21.85 | 26.7 | 26.86 |
| 7 | 7 Feb 2017 | A/Jiangxi/XZS01/2017 | 2107 | 1446 | 25.95 | 33.15 | 33.18 |
| 13 Feb 2017 | A/Jiangxi/XZS02/2017 | 68 | 33 | 28.97 | 32.54 | 32.63 | |
| 9 | 5 June 2017 | A/Hebei/ZLQ01/2017 | 8779 | 1312 | 25.81 | 29.48 | 30 |
| 9 June 2017 | A/Hebei/ZLQ02/2017 | 867 | 265 | 24.22 | 24.45 | 24.9 | |
| 10 June 2017 | A/Hebei/ZLQ03/2017 | 1800 | 693 | 29.41 | 29.29 | 29.56 | |
| 11 June 2017 | A/Hebei/ZLQ04/2017 | 92 | 23 | 32.53 | 31.54 | 31.59 | |
| 11 | 19 Apr 2017 | A/Beijing/DSL01/2017 | 0 | 1654 | 30.8 | None | 33.19 |
| 22 Apr 2017 | A/Beijing/DSL02/2017 | 66 | 2393 | 32.74 | None | 30.27 | |
| 23 Apr 2017 | A/Beijing/DSL03/2017 | 0 | 656 | 35.93 | None | 31.88 | |
aCt values of reverse-transcription polymerase chain reaction (RT-PCR) assay targeting HA, NA 292K, and NA 292R of the H7N9 influenza virus.
bSamples with negative RT-PCR assay results and unavailable Ct values are noted as “None.”
Among 10 other amino acids related to NAI resistance, only the H274Y mutation was detected in 1 sample from case 7; however, this resistant genotype reverted to the WT genotype 6 days later (Table 2). Sustained dual R292K and H274Y mutations were not detected in any of the samples. No mutation was detected at the other 9 sites (Supplementary Table 1).
Mammalian Adaption-Related Mutation in PB2
The PB2 E627K mutation was detected in 18 samples from 7 (77.78%) patients; among these samples, 12 from 5 patients demonstrated coexisting 627K/E (Table 2). In cases 3 and 6, WT 627E was detected in the first sample and disappeared in the second. However, in cases 10 and 11, the WT genotype persisted alongside mutant genotypes. In case 10, the mammalian genotype dominated the viral population, but the WT virus persisted for 9 days, and its proportion only varied slightly over time (Table 2; Figure 2B). In case 11, the mammalian-adapted genotype (627K) predominated in the viral population in the first sample (356K vs 135E), but its predominance was overtaken by the WT genotype 3 days later, at which point 627K constituted only 41.8% of total reads (Table 2). Throughout the 7-day period investigated, the ratio of mutant to WT genotypes fluctuated, and the mammalian-adapted genotype did not reestablish dominance by the final timepoint (Figure 2B).
The D701N substitution was identified in 8 samples from 4 patients, and this mutation coexisted with WT genotypes in all samples (Table 2). Moreover, in 3 patients with multiple timepoint samples, the proportion of the mammalian-adapted genotype (701N) varied slightly during disease development, ranging from 90.08% to 93.55%, 99.44% to 94.53%, and 97.37% to 96.59% in cases 1, 6, and 8, respectively (Table 2). Dual D701N and E627K mutations failed to achieve predominance in any sample. Among other signature amino acids in the PB2 segment, the L89V and A588V mutations were detected in all the samples (Supplementary Table 1), while K526R substitutions occurred in 3 patients (Table 2). The D256G, T271A, M535L, Q591K, and H357N mutations were not detected in these samples (Supplementary Table 1). The coexistence of heterologous amino acids was not detected at these sites.
Signature Amino Acid Variations in Other Segments
Molecular signature sites showed less variation in HA, PB1, PA, and NS1. No multiple basic amino acids were detected in the HA cleavage site, indicating the low pathogenicity of these viruses to poultry (Supplementary Table 1). The T160A mutation occurred in all the samples, with 3 samples containing a mixed T/A population (Table 2). The G186V mutation was detected in 9 patients, potentially favoring the mammalian adaption of avian influenza virus. Additionally, we detected coexisting 186I/V in 4 samples collected serially from case 9 over 6 days. Q226L/I mutations occurred in all patients, and a few prolines (20.9%) were detected at this site in case 2. G228S mutation did not happen in any sample (Supplementary Table 1).
For PB1, viruses in all the samples maintained the WT 99H genotype, but the I368V mutation was present in 9 patients (Supplementary Table 1). V473L and P598L substitutions were not detected in our study, but a sample from case 7 showed the minor occurrence (10.5%) of isoleucine in position 473 (Table 2; Supplementary Table 1). In the PA segment, the mammalian-associated signature K356R and S409N mutations were detected in all available samples, whereas the V100A substitution was not detected (Supplementary Table 1). For NS1, all the samples contained the P42S mutation (Supplementary Table 1), which enhances the pathogenicity of H5N1 avian influenza viruses in mice. Among 3 other signature sites in NS1, which antagonizes type I interferon and tumor necrosis factor α, only N205S was detected, while D92E and G210R mutations were not detected (Supplementary Table 1).
DISCUSSION
Due to the low fidelity of its RNA polymerase, the influenza virus shows high variation and diversity during the development of infection not only at the ecosystem scale but also at the clone level. In this study, we collected sequential samples from H7N9 patients at different timepoints and characterized the genetic variation in the virus during disease development using NGS technology. NA and PB2 exhibited the most dynamic variation, and the majority of signature sites contained both mutant and WT genotypes. Specifically, the signature sites related to antiviral resistance and mammalian adaption underwent dynamic variation during disease progression.
Phylogenetic analysis of HA, NA (Figure 1), and PB2 (Supplementary Figure 1) showed that viral genomes from the same patient at different timepoints clustered together with nearly the same branch length, indicating that viruses from one patient are highly homologous and that intrapatient virus diversity is higher than interpatient diversity. Although several infections in our study occurred in the north of China, the infecting viruses grouped with the eastern virus clade, indicating that the Yangtze Delta remains one of the major sources of human H7N9 virus infection [20].
A previous study showed that NA R292K emerged under antiviral pressure conferred by NAI treatment and was associated with adverse clinical outcomes [12]. This finding led to a comprehensive concern that NAI treatment favors and selects for this drug-resistant genotype and eventually eliminates the WT virus, as occurs with the evolution of antimicrobial-resistant bacteria under antibiotic pressure [21]. However, in cases 7 and 9, NAI-sensitive viruses persisted throughout 6 days of antiviral treatment, and their proportion was only slightly affected. Additionally, in case 9, the viral population already contained substantial numbers of NAI-resistant clones prior to antiviral treatment, indicating that the emergence of the resistant genotype was not triggered by NAI usage (Table 2). Moreover, in cases 2 and 11, the resistant genotype emerged at 1 timepoint and disappeared at the next timepoint, showing that resistant clones were not selected for by NAI pressure. A previous study indicated that the introduction of the R292K mutation into the NA segment leads to competitive fitness loss by avian H7N9 influenza virus [22]. This finding may partially explain why the 292R WT virus persisted simultaneously with resistant 292K under antiviral pressure and the selection of NAI-resistant clones was not as dramatic as that observed in bacteria under antibiotic pressure [21]. Other sites related to NAI resistance showed little variation and were stable during infection. We also validated 292R/K genotypes in our samples using SNP-specific RT-PCR, and most genotypes in the tested samples were confirmed by individual probes (Table 3). However, we found 2 samples demonstrating discordant results between NGS and RT-PCR: a sample collected on 12 February 2017, from case 2 (YB02), and a sample collected on 23 April 2017 from case 11 (DSL02). The 292R/K RT-PCR assay had lower sensitivity than the RT-PCR assay targeting at H7 [19], which may explain the negative results obtained for 292K/R in these samples, as the cycle threshold value for H7 RT-PCR was high and indicated a low viral load. Similar results were also obtained in Wang’s study, in which 4 samples were positive in an H7 RT-PCR assay but negative in an NA 292R/K assay [19]. In DSL02, the ratio of 292K to 292R reads is extremely low, which may explain the negative 292K result obtained for this sample.
The amino acid at residue 627 in PB2 is the most important determinant of host range in influenza A viruses [23, 24]. In cases 3 and 6, this mutation was promoted: the mammalian genotype 627K dominated the viral population, and the WT genotypes were eliminated by the final timepoint. However, the E627K mutation is not always monodirectional, as the WT 627E genotype was maintained alongside the mammalian 627K genotype throughout 10 days of infection in cases 10 and 11, and the proportion of 627K was stable in case 10 (Figure 2B). Moreover, the proportion of 627K mutations underwent fluctuation across 6 timepoints in case 11, and the mammalian genotype failed to achieve consistent predominance throughout 10 days of infection. The D701N mutation was also present in the form of a mixed 701D/N population, and the ratio was constant throughout disease progression (Table 2). Although dual E627K and D701N mutations in the PB2 segment confer higher H7N9 viral polymerase activity and improve viral replication in mammalian cells over a single E627K or D701N mutation [25], this dual mutation did not achieve predominance in any sample during disease development, potentially hampering its capacity for human-to-human transmission. A previous study showed that certain “functionally equivalent” mutations emerged in viruses lacking PB2 627K during the process of human adaption [26]. Our study also detected the PB2 K526R mutation (a “functionally equivalent” mutation) in cases 1 and 2, in whom the viruses lacked PB2 627K (Table 2). However, compensating variants at other sites and segments require further investigation and validation.
Signature amino acid variation in HA, PB1, PA, and NS1 showed less dynamic variation. In case 2, we observed a few reads containing proline at the HA 226 site; this amino acid was detected in only 1 virus among 355 isolates tested from the first 3 waves of human H7N9 infection [20]. In case 8, the isoleucine detected at the first timepoint transformed to leucine after 4 days and persisted thereafter, indicating that isoleucine may be an intermediate amino acid during the transition from glutamine to leucine at the 226 position. Other molecular signatures, such as T160A and G186V/I mutations, were detected in some patients, but only a few samples contained mixed genotypes. Molecular signatures in PB1, PA, and NS1 showed less variation, similar to that of HA.
Overall, whole H7N9 genomes were obtained from 23 samples (79.3%) in our study, and the majority of viral segments were fully covered in the other 6 samples. Thus, all known molecular markers across the H7N9 genome were likely detected. However, there are several limitations to our study. First, the patient sample size in our study was small and insufficient to establish relationships between variations in signature markers and patients’ clinical indices. Second, direct amplification of the complete viral genome from clinical samples is more difficult than that from clinical isolates; therefore, sequence information was missing for some timepoints, and the number of patients with data from multiple samples was small. Last, most patients were sampled after hospital admission, when some time had already passed from disease onset. Thus, some strong selective changes occurring during the early stage of infection may have been missed in this study. However, the sequential sample data presented here clearly indicated that dynamic variation and reversion happened in the H7N9 patients in vivo.
In conclusion, the H7N9 virus underwent dynamic evolution during infection in certain patients, and the amino acids at NA 292 and PB2 627 were “hot spot” molecular markers exhibiting the most vigorous variation. Even under antiviral pressure conferred by NAI and the emergence of drug-resistant mutation NA R292K, the WT 292R virus persisted in viral populations, and selection for resistant genotypes was not obligatory. Similarly, although the PB2 E627K and D701N mutations favor avian influenza virus infection in mammals, they undergo fluctuation and reversion during mammalian adaption. Therefore, the variation of H7N9 virus should be monitored at the quasispecies level to clarify its pathogenicity during disease development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We acknowledge the contributions of the technical staff of the Laboratory of Clinical Microbiology and Infectious Diseases, China–Japan Friendship Hospital. We thank Lei Yang and Dr Weifei Zhu at the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, for their help with viral phylogenetic analysis.
Financial support. This work was supported by the National Science Fund for Distinguished Young Scholars (grant number 81425001/H0104 to B. C.); the National Key Technology Support Program from the Ministry of Science and Technology (grant number 2015BAI12B11 to B. C.); and the 13th Five-Year Jiangsu Province Health, Science and Technology Key Project (grant number ZDRCA2016046 Q. G.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gao R, Cao B, Hu Y, et al. . Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888–97. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Q, Shi J, Deng G, et al. . H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 2013; 341:410–4. [DOI] [PubMed] [Google Scholar]
- 3. Ke C, Mok CKP, Zhu W, et al. . Human infection with highly pathogenic avian influenza A(H7N9) virus, China. Emerg Infect Dis 2017; 23:1332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang L, Zhu W, Li X, et al. . Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in Mainland China. J Virol 2017; 91:e01277–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Monthly risk assessment summaryhttp://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/. Accessed 1 February 2018.
- 6. Watanabe T, Kiso M, Fukuyama S, et al. . Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013; 501:551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou J, Wang D, Gao R, et al. . Biological features of novel avian influenza A (H7N9) virus. Nature 2013; 499:500–3. [DOI] [PubMed] [Google Scholar]
- 8. Zhu H, Wang D, Kelvin DJ, et al. . Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 2013; 341:183–6. [DOI] [PubMed] [Google Scholar]
- 9. Wang D, Yang L, Gao R, et al. . Genetic tuning of the novel avian influenza A(H7N9) virus during interspecies transmission, China, 2013. Euro Surveill 2014; 19. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Li X, Guo J, et al. . The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J Gen Virol 2014; 95:779–86. [DOI] [PubMed] [Google Scholar]
- 11. Marjuki H, Mishin VP, Chesnokov AP, et al. . Neuraminidase mutations conferring resistance to oseltamivir in influenza A(H7N9) viruses. J Virol 2015; 89:5419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y, Lu S, Song Z, et al. . Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 2013; 381:2273–9. [DOI] [PubMed] [Google Scholar]
- 13. Sha J, Chen X, Ren Y, et al. . Differences in the epidemiology and virology of mild, severe and fatal human infections with avian influenza A (H7N9) virus. Arch Virol 2016; 161:1239–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seifert D, Beerenwinkel N. Estimating fitness of viral quasispecies from next-generation sequencing data. Curr Top Microbiol Immunol 2016; 392:181–200. [DOI] [PubMed] [Google Scholar]
- 15. Jonges M, Welkers MR, Jeeninga RE, et al. . Emergence of the virulence-associated PB2 E627K substitution in a fatal human case of highly pathogenic avian influenza virus A(H7N7) infection as determined by Illumina ultra-deep sequencing. J Virol 2014; 88:1694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou XH, Chen WB, Zhao X, et al. . Evaluation of a single-reaction method for whole genome sequencing of influenza A virus using next generation sequencing. Biomed Environ Sci 2016; 29:41–6. [DOI] [PubMed] [Google Scholar]
- 17. Zhou B, Donnelly ME, Scholes DT, et al. . Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J Virol 2009; 83:10309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang W, Song Z, Guan W, et al. . PCR for detection of oseltamivir resistance mutation in influenza A(H7N9) virus. Emerg Infect Dis 2014; 20:847–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang D, Yang L, Zhu W, et al. . Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol 2016; 90:5561–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science 2017; 355:826–30. [DOI] [PubMed] [Google Scholar]
- 22. Yen HL, Zhou J, Choy KT, et al. . The R292K mutation that confers resistance to neuraminidase inhibitors leads to competitive fitness loss of A/Shanghai/1/2013 (H7N9) influenza virus in ferrets. J Infect Dis 2014; 210:1900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mok CK, Lee HH, Lestra M, et al. . Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 2014; 88:3568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamayoshi S, Fukuyama S, Yamada S, et al. . Amino acids substitutions in the PB2 protein of H7N9 influenza A viruses are important for virulence in mammalian hosts. Sci Rep 2015; 5:8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu W, Li L, Yan Z, et al. . Dual E627K and D701N mutations in the PB2 protein of A(H7N9) influenza virus increased its virulence in mammalian models. Sci Rep 2015; 5:14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mänz B, de Graaf M, Mögling R, et al. . Multiple natural substitutions in avian influenza A virus PB2 facilitate efficient replication in human cells. J Virol 2016; 90:5928–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.