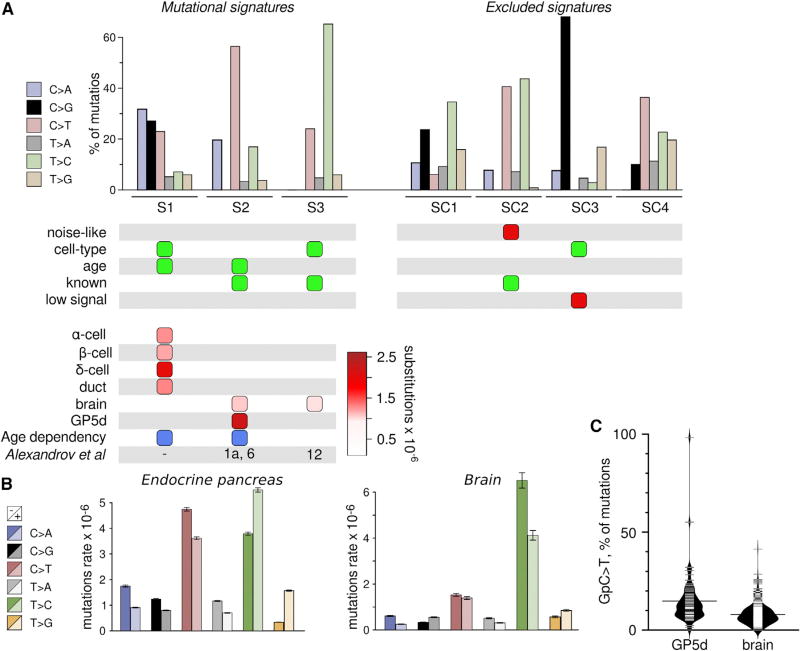
Figure 4. Mutational Signatures Derived from scRNA-Seq Data.
(A) Single-nucleotide substitutions in 3,003 cells from pancreas, brain, and the colon cancer cell line GP5d were organized into mutational signatures using non-negative matrix factorization followed by agglomerative hierarchical clustering. Bar plot illustrates the percent of mutations attributed to each substitution type in each of the three signatures (S1–S3, left) and the four excluded signatures (SC1–SC4, right). Colors as in (A). Panel below the bar plot indicates selection items for determining whether to exclude the signature. Green, cause for inclusion; red, cause for exclusion. Bottom panel denotes the presence of a signature (columns) in a cell type (rows), with color scale indicating strength of signature as median substitution rate for cells of the indicated type. Blue boxes denote significant association between signature load and donor age. Bottom row indicates equivalent signatures from Alexandrov et al. (2013b).
(B) Strand specificity differs between cell types. Mutations were annotated based on whether the mutated pyrimidine occurred on the transcribed (−) or untranscribed (+) strand. Bars represent mean ± SEM of raw substitution counts in endocrine cells (left) and brain cells (right). Note that endocrine cells have a strong strand bias for the transcribed strand for C > A, C > G, and C > T substitutions (p = 1.00E-79, 1.37e-28, and 6.40E-34, respectively; Wilcoxon test, n = 1,429) previously observed in oxidative stress-related tumor signatures, while brain has a bias for T > C substitutions on the transcribed strand (p = 3.41E-11; Wilcoxon test, n = 466) similar to tumor signature 12 (Alexandrov et al., 2013b).
(C) Signature S2 is composed of two sub-signatures corresponding to cancer signatures 1 and 6. Violin plot show C > T substitutions with a preceding G as a fraction of all substitutions in a cell, which is a hallmark of cancer signature 6 and that separates GP5d and brain cells (p = 7.156E-11; Wilcoxon test, n = 73 for GP5d and n = 332 for brain cells).