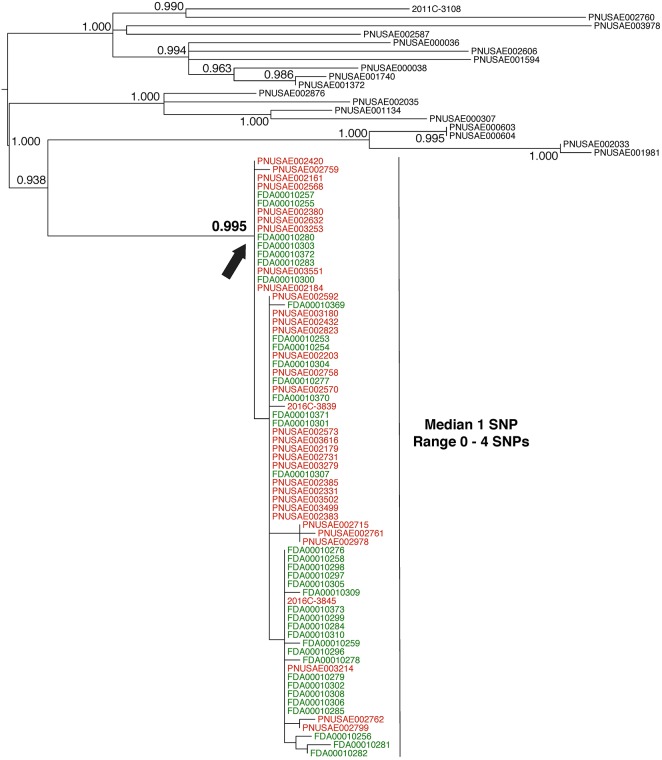
FIGURE 3.
Phylogenetic analysis of genome sequences obtained from Escherichia coli isolates implicated in a 2016 flour outbreak. A SNP matrix was generated for 89 isolates with the CFSAN SNP pipeline v0.8.0 (Davis et al., 2015). The SNP matrix was phylogenetically analyzed with GARLI v2.01.1067 (Zwicki, 2006), using the K80 and HKY nucleotide substitution models. Branch labels indicate support values for 1000 bootstrap replicates. Bootstrap values less than 0.800 are not shown. Red labels indicate clinical isolates and green labels indicate flour isolates related to the outbreak. Black labels indicate isolates that were determined by epidemiology and traceback investigations to not be involved in the outbreak. The black arrow identifies the node that represents the last common ancestor of isolates included in the 2016 outbreak.