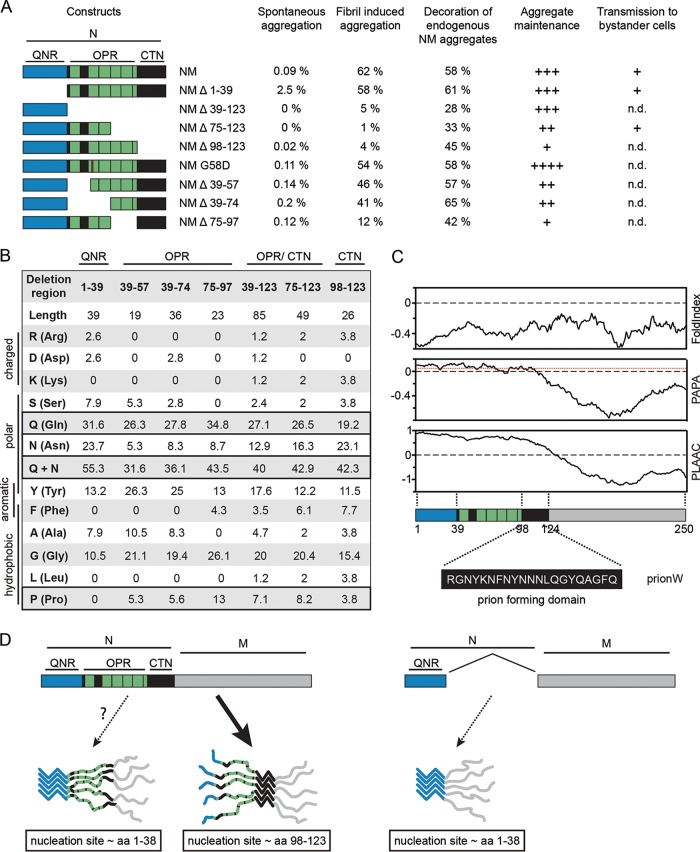
FIG 9.
Carboxy-terminal OPR and the CTN are involved in nucleation, template-assisted aggregation, and prion maintenance. (A) Summary of results. Spontaneous aggregation, template-assisted aggregation, maintenance, and transmission to bystander cells by bulk cell populations or cell clones stably expressing the respective NM-HA proteins. Coaggregation with endogenous NM-GFP aggregates was determined following transient overexpression of wild-type and mutant NM-HA in N2a NM-GFPagg cells. Shown is the percentage of cells showing NM-GFP/NM-HA aggregate colocalization per NM-GFP/NM-HA aggregate-bearing cell. Aggregate maintenance was assessed as a proxy for seed multiplication: ++++, slightly increased mitotic stability compared to NM-HAagg control cells (fold change, >−0.5 and <0); +++, only slightly altered mitotic stability compared to NM-HAagg control (fold change, >0 and <0.5); ++, lower mitotic stability compared to control (fold change, >0.5 and <1); +, lowest mitotic stability (fold change, >1 and <2). For transmission to bystander cells, a plus sign indicates that at least 1 in 1,000 recipient cells contained NM-GFPagg. n.d., not done. (B) Amino acid composition of different subdomains of the N domain. The position of the region within NM and its length are indicated at the top. The abundances of specific residues within the region are given as percentages. (C) Prediction of disorder (FoldIndex [86]) and prion domains according to PAPA (16), PLAAC (15), or pWaltz/PrionW (80). Images were designed using PLAAC (http://plaac.wi.mit.edu/). The region identified as the prion nucleating domain by PrionW is shown at the bottom. This region falls within the CTN (aa 98 to 123). (D) Putative model of NM prion nucleation and seeding in N2a cells. NM contains two putative nucleation sites. Within the mammalian cytosol, the carboxy-terminal site comprising aa residues 98 to 123 preferentially drives nucleation and template-assisted seeding (left). The amino-terminal nucleation site (residues 1 to 38) likely mediates self-recognition and assembly when the carboxy-terminal site is deleted (right).