FIGURE 2.
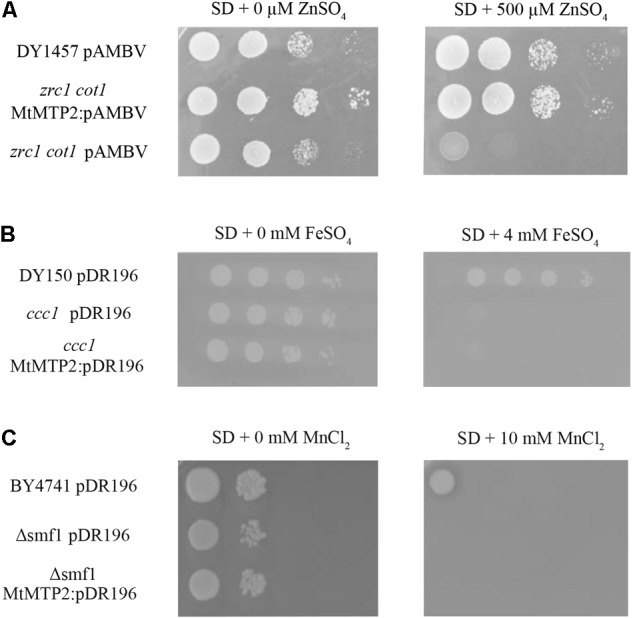
MtMTP2 yeast complementation assays. (A) Yeast strain DY1457 was transformed with the pYPGE15 empty vector, while the double mutant zrt1/cot1 was transformed with the empty pYPGE15 or with pYPEG15 containing MtMTP2 coding DNA sequence. Serial dilutions (10×) of each transformant were grown for 3 days at 28°C on SD media with all the required amino acids and 500 μM ZnSO4. Positive control was obtained without supplementing the media with ZnSO4. (B) Yeast strain DY150 was transformed with the pDR196 empty vector, while ccc1 mutant was transformed either with the empty pDR196 or with pDR196 containing MtMTP2 coding DNA sequence. Serial dilutions (10×) of each transformant were grown for 3 days at 28°C on SD media with all the required amino acids and 4 mM FeSO4. Positive control was obtained without supplementing the media with FeSO4. (C) Yeast strain BY4741 was transformed with the pDR196 empty vector, while a deletion strain in smf1 was transformed either with empty pDR196 or with pDR196 containing MtMTP2 coding DNA sequence. Serial dilutions (10×) of each transformant were grown for 3 days at 28°C on SD media with all the required amino acids and 10 μM MnCl2. Positive control was obtained without MnCl2 supplementation.
