FIGURE 2.
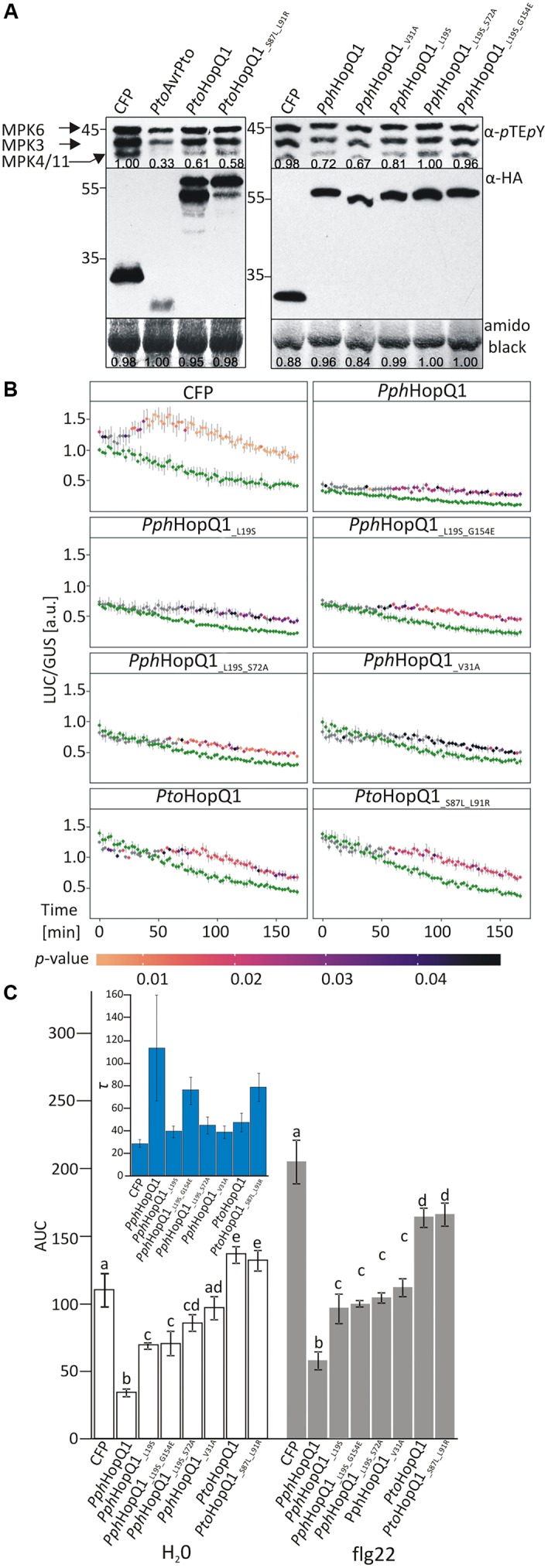
Effect of HopQ1 variants’ expression on defense-related MAPK activation. (A) Arabidopsis protoplasts were transformed with plasmids encoding hemagglutinin (HA)-tagged HopQ1 variants under the control of the cauliflower mosaic virus 35S promoter. Expression of cyan fluorescent protein (CFP) and AvrPto served as negative and positive controls, respectively. Fourteen hours after transformation, protoplasts were treated with 100 nM flg22 for 10 min. MAPK activation was monitored by immunoblot analysis with anti-pTEpY antibodies and the expression level of HopQ1 variants was checked with anti-HA antibodies. Amido black staining of the membranes was used to demonstrate equal loading. The numbers correspond to ImageJ-based quantification of the protein band intensities (MAPK activation strength is the sum of all three MAPK bands). (B) Arabidopsis protoplasts were co-transformed with constructs expressing HopQ1 variants, pNHL10-LUC (luciferase) as a reporter and pUBQ10-GUS (β-glucuronidase). Luciferase activity was recorded for 3 h, following flg22 treatment, and depicted as LUC/GUS ratios. Data for each protein variant were analyzed using repeated measures ANOVA, yielding significant effects of variant, time and their interaction (p < 0.001). Differences between H2O-treated samples (green traces) and flg22 treatments were tested with Student’s t-test. Statistical significant differences in the flg22-treated samples as compared to the H2O-treated samples are highlighted by the color-coded p-values adjusted using Benjamini–Hochberg procedure. (C) Area under the curve (AUC) values were calculated for the graphs. One-way ANOVA was performed separately for both treatments and was followed by Tukey HSD post hoc test. Letters correspond to statistically homogenous groups (p < 0.05). Inlet: τ parameter values obtained after curve fitting to the fold changes for each protein variant (see Supplementary Figure 1). Bars correspond to standard errors in parameter estimation. The experiment was performed three times with similar results.
