FIGURE 2.
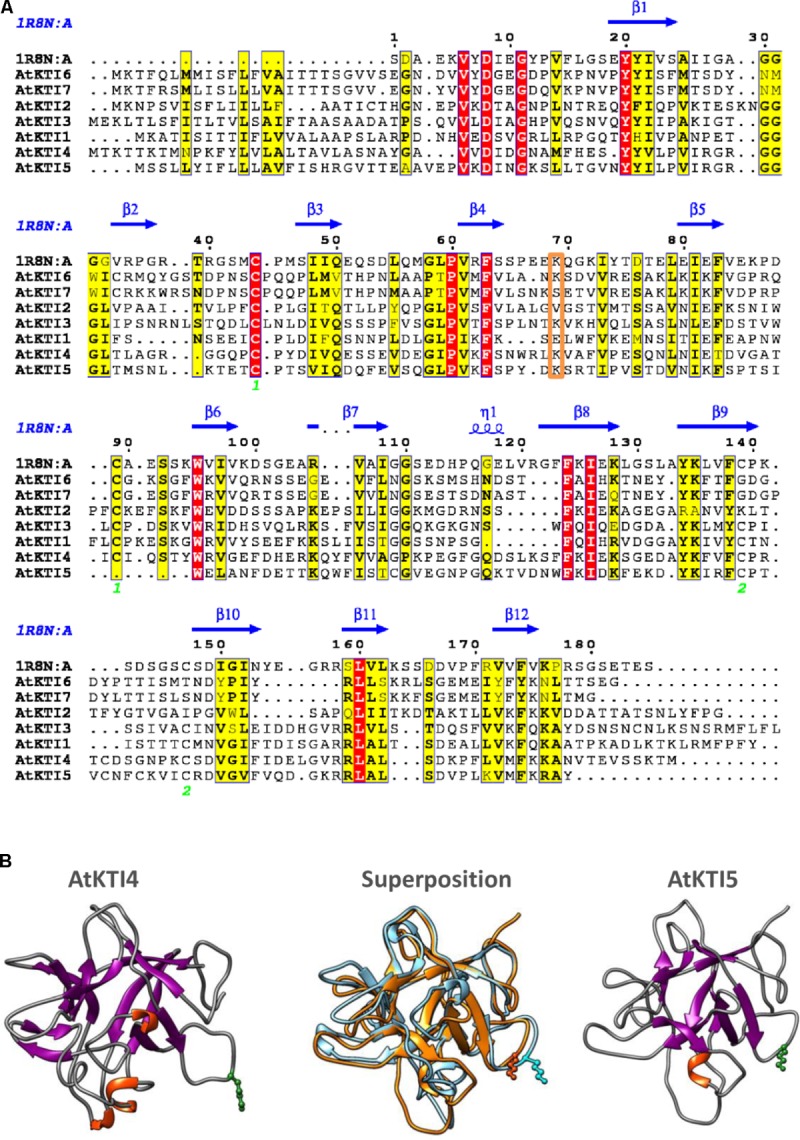
Sequence-structure analysis of Kunitz-type inhibitors. (A) Multiple sequence alignment of Arabidopsis KTIs. The Kunitz type inhibitor of Delonix regia (PDB ID 1R8N) was included in the alignment to infer secondary structure locations. Conserved residues are in red boxes. Similar residues are indicated in black bold characters and boxed in yellow. Green numbers at the bottom indicate disulphide bridge topology. Putative reactive site is boxed in orange. The figure was made with the ESPript 3.0 web server. (B) Ribbon diagrams showing structural models for AtKTI4 and AtKTI5, and their superposition (blue, AtKTI4; orange, AtKTI5). Conserved reactive Lys residue is colored in green. Modelization and visualization were made by SWISS-Model and Chimera tools.
