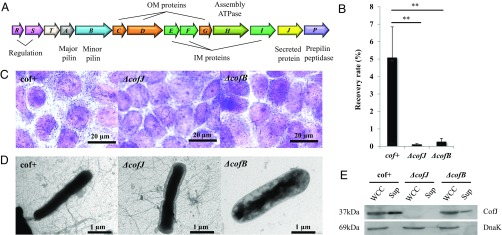
Fig. 1.
CFA/III-mediated E. coli adherence to Caco-2 cells. (A) Genetic organization of the cof gene cluster. Predicted functions for each gene product are indicated at the Top and Bottom of the figure. (B) Adherence values correspond to the recovery rate after incubation for 3 h. cof+ is an E. coli strain HB101 harboring all cof operon genes on the plasmid pTT240, ΔcofJ is a cofJ deletion mutant, and ΔcofB is a cofB deletion mutant. The experiments were performed five times. cof+, ΔcofJ, and ΔcofB recovery rates were 5.08 ± 1.76%, 0.12 ± 0.04%, and 0.27 ± 0.19%, respectively. **P < 0.005 vs. cof+. (C) Micrographs showing adhesion of cof+, ΔcofJ, and ΔcofB strains to Caco-2 cells. (D) TEM images of cof+, ΔcofJ, and ΔcofB strains grown on CFA agar plates. (E) Western blot analysis of whole cell culture (WCC, Left) and supernatant (Sup, Right) of cof+, ΔcofJ, and ΔcofB strains using anti-CofJ antibody. DnaK was detected with anti-DnaK antibody as a loading control.