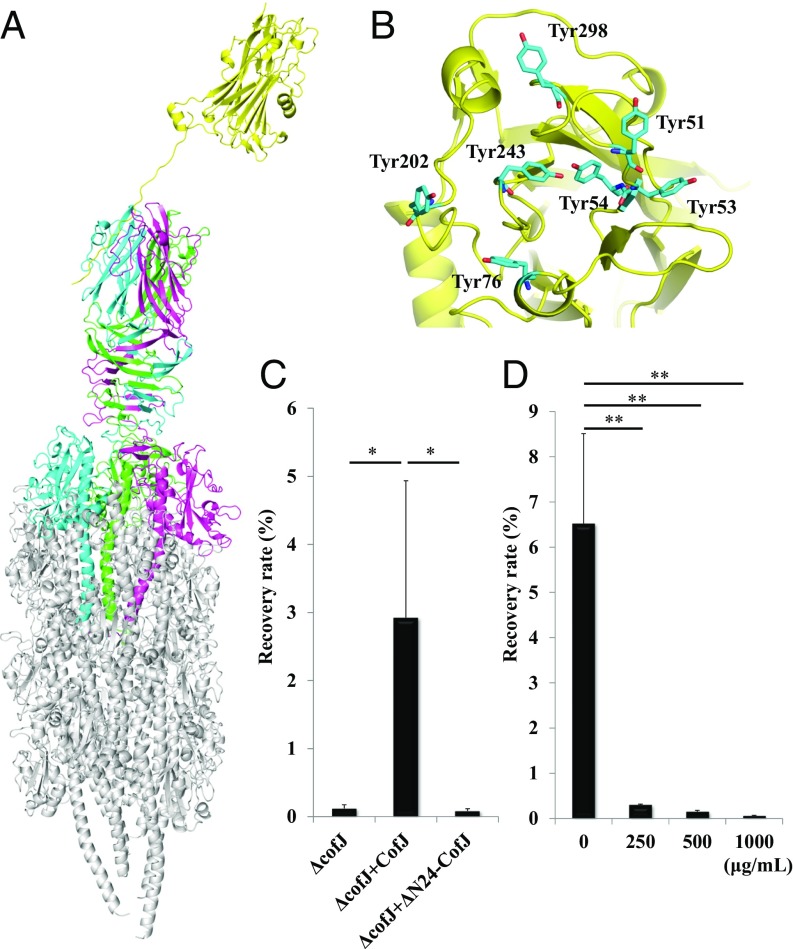
Fig. 4.
Structural model of the CofJ–CFA/III pilus complex. (A) Side view of the CofJ–CFA/III pilus depicted as a ribbon model, built by superimposing the CofJ (1–24)–CofB crystal structure onto our previously reported CFA/III pilus model with PyMOL. The CofJ N-terminal disordered residues (Ser1 to Ser4 and Lys16 to Asp22) were modeled and connected with the CofJ globular domain situated above the CofB homotrimer by using the program Coot (52). The CofJ monomer (yellow) is situated further above the minor pilin CofB homotrimer (cyan, magenta, and green) at the major pilin CofA (gray) pilus tip. (B) Ribbon representation of CofJ shown in the Top region with seven tyrosine residues (cyan stick representation). (C) Adherence values correspond to recovery rates after 3-h incubation. ΔcofJ is the cofJ deletion mutant. Experiments were performed five times. ΔcofJ alone, ΔcofJ plus CofJ, and ΔcofJ plus ΔN24-CofJ recovery rates were 0.12 ± 0.05%, 2.92 ± 2.01%, and 0.08 ± 0.03%, respectively. *P < 0.05 vs. ΔcofJ mixed with CofJ. (D) Results of cof+ adherence inhibition assay using anti-CofJ polyclonal antibody Fab fragments. Adherence values are as in C. Bottom values are anti-CofJ IgG antibody Fab fragment concentration. Experiments were performed five times. cof+ alone, cof+ plus 250, 500, and 1,000 µg/mL Fab fragment recovery rates were 6.53 ± 1.98%, 0.30 ± 0.01%, 0.15 ± 0.03%, and 0.06 ± 0.01%. **P < 0.005 vs. cof+ alone.