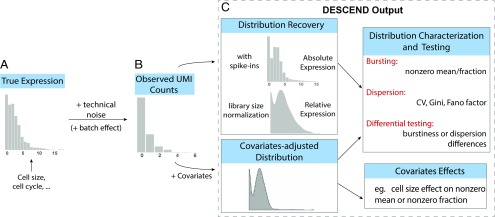
Fig. 1.
Illustration of the framework. (A and B) The cross-cell distribution of observed counts (B) is assumed to be a convolution of the distribution of true gene expression (A) and technical noise. (C) For each gene, the output of DESCEND includes the distribution of the absolute expression levels when spike-ins are available, the distribution of relative expression with library size normalization, the distribution of covariates-adjusted expression level if covariates are presented, estimates of the bursting and dispersion parameters, differential testing results comparing the change between two cell populations, and the effects of observed covariates on gene expression.