Fig. 7.
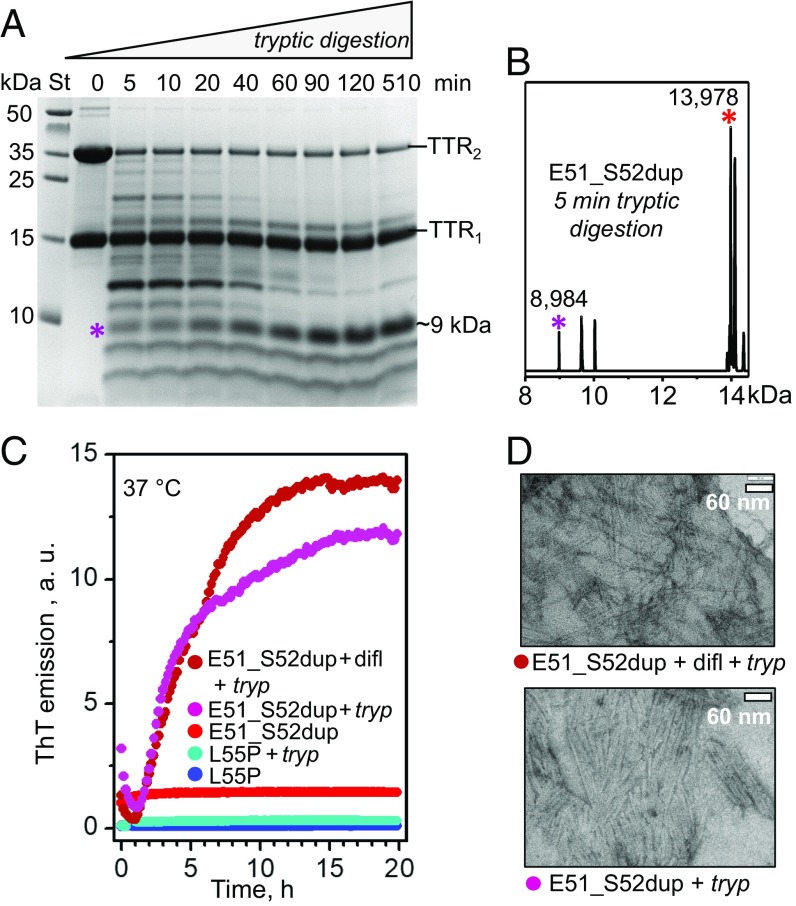
Proteolysis of Glu51_Ser52dup by trypsin leads to amyloid formation. (A) SDS/PAGE of Glu51_Ser52dup at various stages of limited proteolysis. The protein (1 mg/mL) was incubated with trypsin at 37 °C, 200:1 substrate/enzyme weight ratio, for 5–510 min as indicated on the lanes; 0 indicates intact protein before incubation. An asterisk indicates the ∼9-kDa fragment that is formed after 5 min and gradually accumulates over time. St, molecular weight standards. (B) Electrospray ionization mass spectrometry of the incubation mixture at 5 min from A reveals a C-terminal fragment with molecular mass of 8,984 Da corresponding to residues 49–129 of Glu51_Ser52dup, along with the full-length duplication mutant (residues 1–129, 13,978 Da). (C) Amyloid formation upon limited proteolysis of mutant TTR monitored by ThT emission. Glu51_Ser52dup (warm colors) and Leu55Pro (cold colors) were incubated at 37 °C alone, with trypsin (1 mg/mL TTR, 200:1 weight ratio of TTR monomer to enzyme), or with trypsin and diflunisal [25:1 (mol/mol) drug/TTR monomer, 1.8 mM diflunisal]. Only samples containing E51_S52dup and trypsin, either with or without diflunisal, showed ThT binding to amyloid-like structures. (D) Transmission electron micrographs of negatively stained samples after incubation with trypsin for 5 d with double-orbital shaking at 168 rpm, 37 °C in a TECAN fluorescence microplate reader (Fisher Scientific). Samples containing Glu51_Ser52dup and trypsin, either with (Top, 25:1 drug/TTR monomer molar ratio) or without (Bottom) diflunisal, showed massive fibril formation, while other samples showed few fibrils. difl, diflunisal; tryp, trypsin.