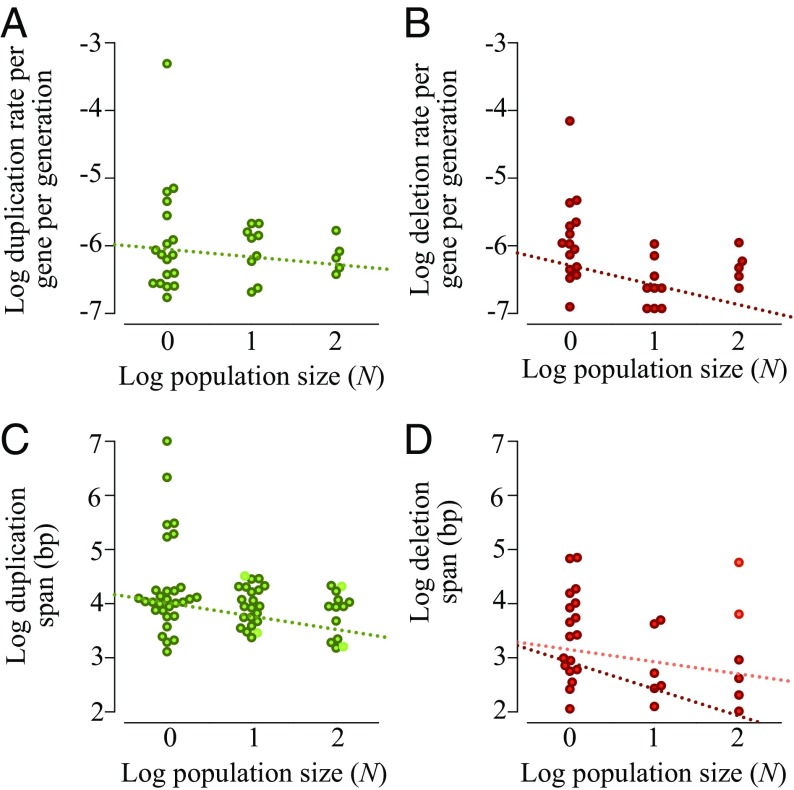
Fig. 1.
Linear regression showing the rate of accumulation and span of copy-number changes as a function of population size. (A) No significant effect of population size on the accumulation of gene duplications (Kendall’s τ = −0.02, P = 0.87; Kruskal–Wallis H = 0.11, P = 0.95). (B) Significant negative association between population size and the accumulation of gene deletions (Kendall’s τ = −0.35, P = 0.02; Kruskal–Wallis H = 9.95, P = 0.01). (C) Negative correlation between duplication span and population size. The Pearson correlation for all unique duplications (including polymorphic) is −0.28 (P = 0.02; permutation 95% CI: −0.23, 0.25, P = 0.007). (D) No significant correlation between deletion spans of single-copy DNA and population size (r = −0.21, P = 0.25; permutation CI: −0.35, 0.36, P = 0.12, hatched orange line). There is a significant correlation between the span of fixed deletions and population size [r = −0.46, P = 0.01; permutation CI: (−0.37, 0.38), P = 0.005, hatched maroon line]. Fixed and polymorphic copy-number changes in C and D are indicated by darker and lighter filled circles, respectively.