Figure 3. apoEVs are enriched with spliceosomal proteins and U snRNAs.
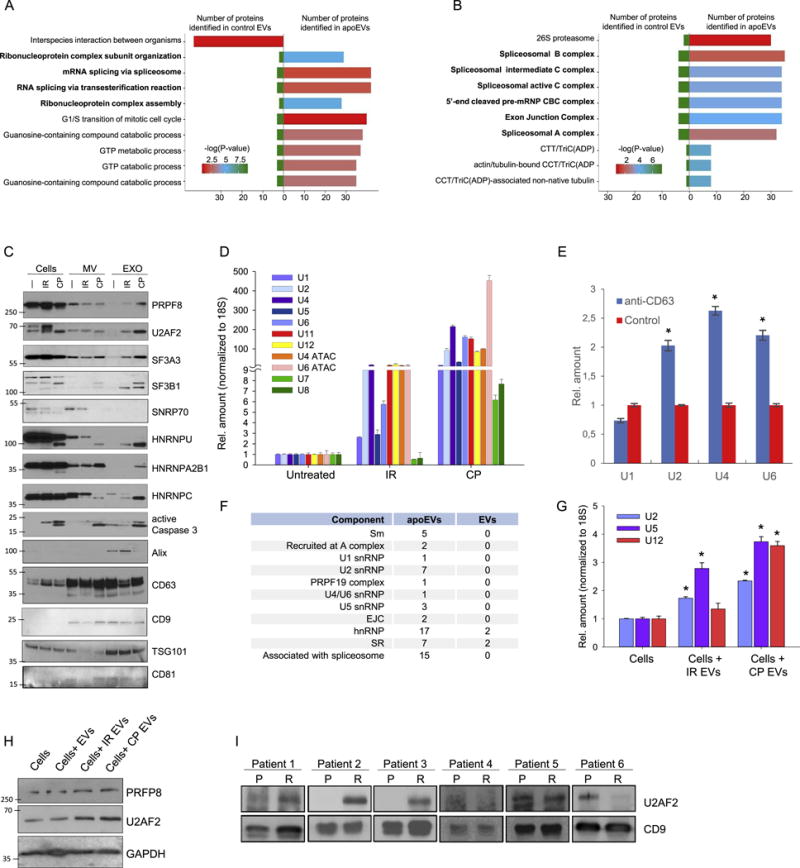
(A and B) Analysis of proteins identified by LC-MS/MS in apoEVs and control EVs with Gene Ontology Biological process database (A) or National Cancer Institute database (B).
(C) Western blotting analysis of exosome-like vesicles (EXO) and microvesicles (MV) purified from GBM157 spheres that were either untreated (—), lethally irradiated (IR), or treated with cisplatin (CP); PRPF8, U2AF2, SF3A3, SF3B1, SNRNP70, HNRNPU, HNRNPA2B1, HNRNPC – splicing factors; active Caspase 3 – apoptosis marker, Alix, CD63, CD9, TSG101, CD81 – exosome markers.
(D) qRT-PCR analysis of U snRNAs in vesicles purified as in “C”.
(E) qRT-PCR analysis of U snRNAs in vesicles purified by immunoprecipitation with anti-CD63 or control antibodies.
(F) Number of proteins related to different spliceosomal components that were identified in apoEVs and control EVs.
(G) qRT-PCR analysis of U snRNAs in GBM157 cells incubated for 10 hr with apoEVs from lethally irradiated or cisplatin treated cells, untreated cells were used as a control.
(H) Western blotting analysis of GBM157 neurospheres incubated with EVs from untreated cells and apoEVs purified from lethally irradiated or CP treated cells for 16 hr, untreated cells were used as a control.
(I) Western blotting analysis of exosomes isolated from blood serum samples obtained from GBM patients before (P) and after (R) post-surgical chemo- and radio-therapies. CD9 was used as a loading control.
All quantitative data are average ± SD; *p < 0.01; See also Figure S4 and Table S1.
