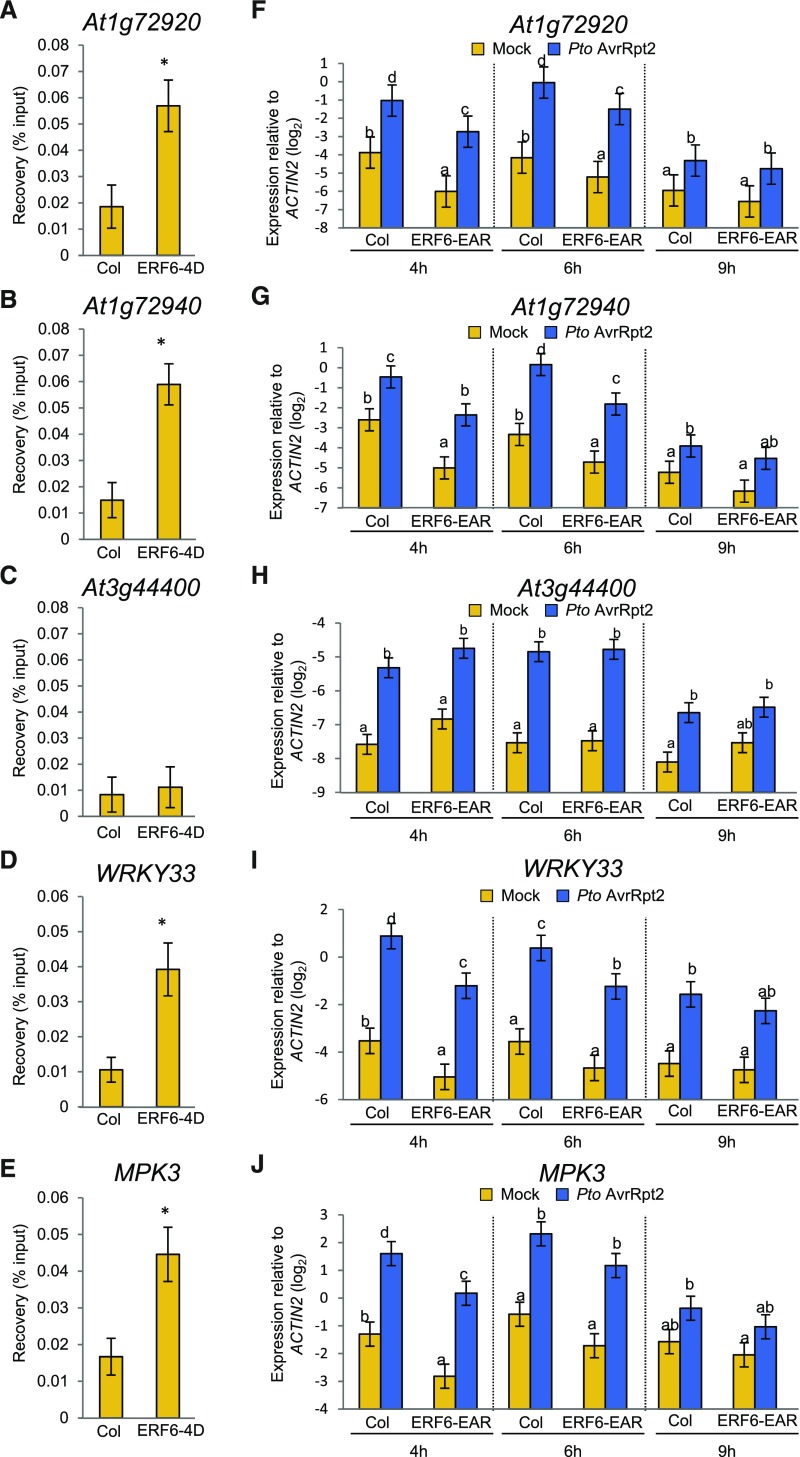
Figure 12.
ERF6 Regulates Expression of TIR-NB-LRRs, WRKY33, and MPK3 and Binds to GCC Box in the Promoters.
(A) to (E) ChIP-qPCR was performed using the ERF6-4D line. The input and ChIP DNA was analyzed by qPCR using primers spanning the GCCGCC sequence in the promoters of At1g72920 (A), At1g72940 (B), At3g44400 (C), WRKY33 (D), and MPK3 (E). See also Supplemental Data Set 12. Bars represent means and standard errors of the percentage of input values of the ChIP DNA, calculated from three independent experiments. Asterisks indicate statistically significant differences compared with Col-0 (Col) (P < 0.05, two-tailed t tests).
(F) to (J) Leaves of Col-0 (Col) and ERF6-EAR plants were infiltrated with mock or Pto AvrRpt2 (OD600 = 0.001) and were harvested at 4, 6, and 9 hpi. The expression levels of At1g72920 (F), At1g72940 (G), At3g44400 (H), WRKY33 (I), and MPK3 (J) were measured by RT-qPCR. Bars represent means and standard errors of two independent experiments. The Benjamini-Hochberg method was used to adjust P values to correct for multiple hypothesis testing. Statistically significant differences are indicated by different letters at each time point (adjusted P < 0.05).