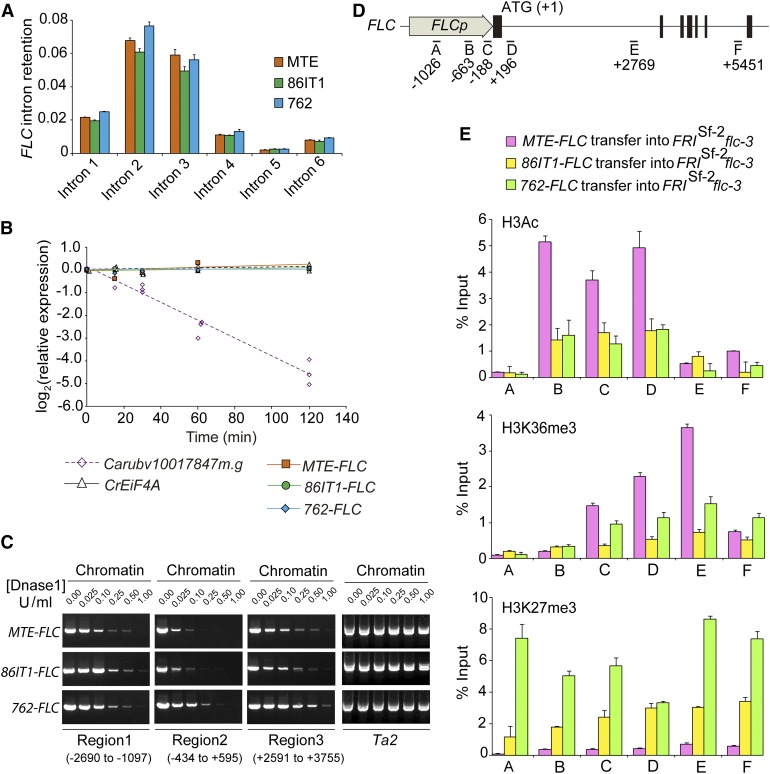
Figure 4.
Sequence Deletion in the FLC 5′ UTR Is Correlated with Altered Chromatin Structure State and Histone Modifications.
(A) FLC splicing efficiency in three C. rubella accessions. Intron retention was calculated as the ratio of unspliced to total (spliced + unspliced) transcripts for all six FLC introns. Results were normalized to C. rubella TUBULIN (Carubv10017235m) results and are shown as means ± sd (n = 3 biological replicates; with 15 seedlings per replicate).
(B) RNA stability assay in three C. rubella accessions. After the addition of cordycepin, the rate of mRNA degradation was measured by RT-qPCR to determine the level of remaining transcripts relative to that of an untreated sample. C. rubella EIF4A and Carubv10017847m.g were used as controls, representing a stable and an unstable transcript, respectively.
(C) Sensitivity of three regions (regions 1 to 3) of C. rubella FLC to increasing dosages of mononuclease in three transgenic plants. Chromatin with loose nucleosome occupation is more susceptible to mononuclease digestion and is inefficiently amplified by PCR. The A. thaliana Ta2 region in three transgenic plants was used as an insensitive control (Shu et al., 2013). The position of each region is labeled, and the A of ATG is referred to as +1.
(D) Regions of C. rubella FLC used for ChIP-qPCR assays.
(E) ChIP-qPCR analyses of six regions of C. rubella FLC. Data are from pooled T2 transgenic plants using antibodies against H3Ac (top), H3K36me3 (middle), and H3K27me3 (bottom), respectively. ChIP values were normalized to their respective DNA inputs. Data are shown are the means ± sd (n = 3 biological replicates; each replicate was obtained from at least 3 g of tissue corresponding to roughly 500 10-d-old seedlings from 10 independent T2 transgenic lines).