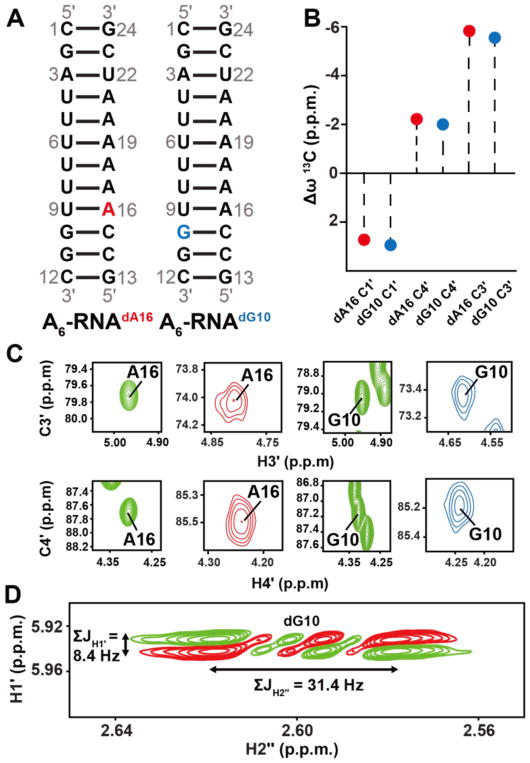
Figure 2.
13C chemical shifts in C3′-endo deoxyribose. (A) A6-RNA duplex highlighting the dA16 and dG10 dexoyribose substitutions. (B) Differences (Δω = ωA6RNA/dNTP – ωA6DNA) between sugar13C chemical shifts at A16 and G10 when comparing A6-DNA duplex versus dA16 and dG10 substituted A6-RNA. (C) 2D C3′-H3′ and C4′-H4′ HSQC spectra of the corresponding A6-DNA duplex (green) and dA16 (red) and dG10 (blue) substituted A6-RNA. (D) The sum of the scalar coupling constants ΣJH1′ and ΣJH2″ in DQF-COSY77 indicate that the sugar pucker at dA16 and dG10 in substituted A6-RNA is C3′-endo42,68.