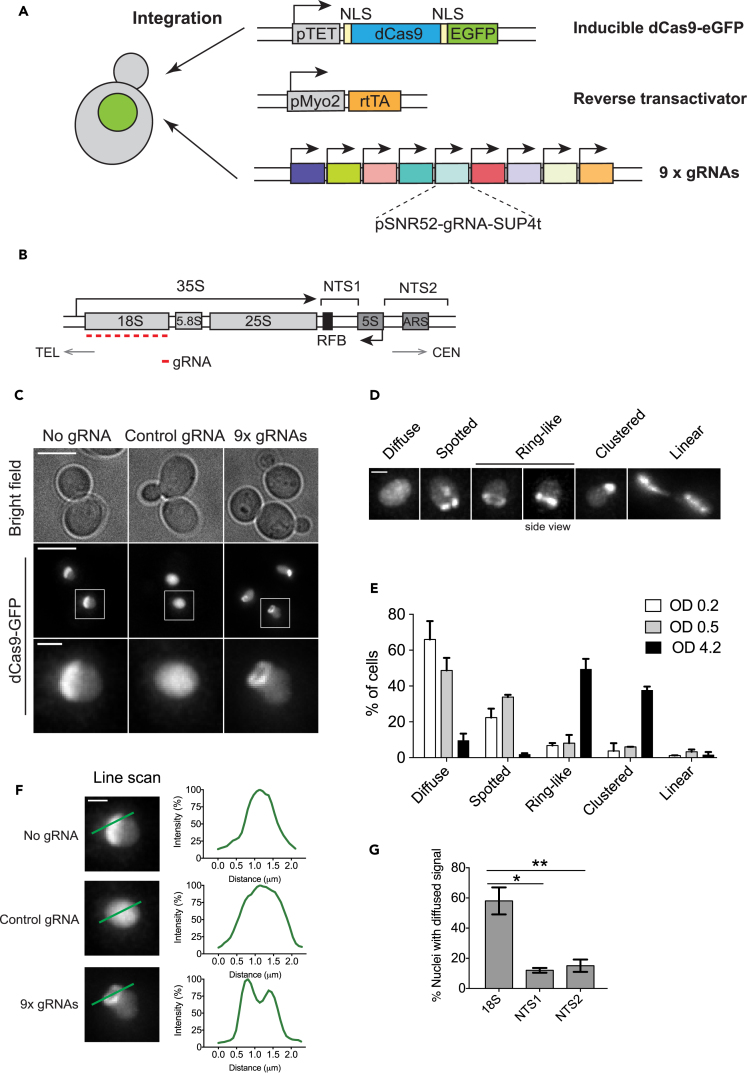
Figure 1.
CRISPR Live Imaging System for Visualizing the Budding Yeast rDNA
(A) CRISPR imaging platform for the budding yeast. Doxycycline-inducible dCas9-GFP, rtTA, and nine gRNAs that target the 18S rDNA locus were stably integrated into the genome.
(B) A single ∼9.1-kb rDNA repeat unit of the budding yeast. The orientation toward telomere (TEL) and centromere (CEN) and the direction of rRNA transcription are indicated. Nine rDNA-targeting gRNAs were designed to bind the template strand of the 18S rDNA (red bars). The location of the replication fork barrier (RFB) and two non-transcribed spacer (NTS) regions are indicated.
(C) CRISPR imaging of the rDNA chromatin. Deconvolved, maximum projected z series are shown. Scale bar, 5 μm (overview) and 1 μm (inset). The control gRNA does not target the yeast genome.
(D) Classification of rDNA phenotypes by CRISPR imaging. For simplicity, only nuclei are shown. Scale bar, 1 μm.
(E) Distribution of rDNA phenotypic classes in cells grown to different densities. n = 3; error bars, mean ± SEM.
(F) Line scan analysis for different dCas9-GFP signals. Fluorescence intensity was normalized to the peak intensity value. Scale bar, 1 μm.
(G) Quantification of cells showing diffuse dCas9-GFP signal. dCas9-GFP-expressing cells carry genome-integrated gRNAs targeting either the 18S rDNA or the non-transcribed spacers NTS1 or NTS2. n = 3; error bars, mean ± SEM. p Values were calculated by unpaired Student's t test. *p < 0.05, **p < 0.01.
See also Figures S1–S3 and S7.