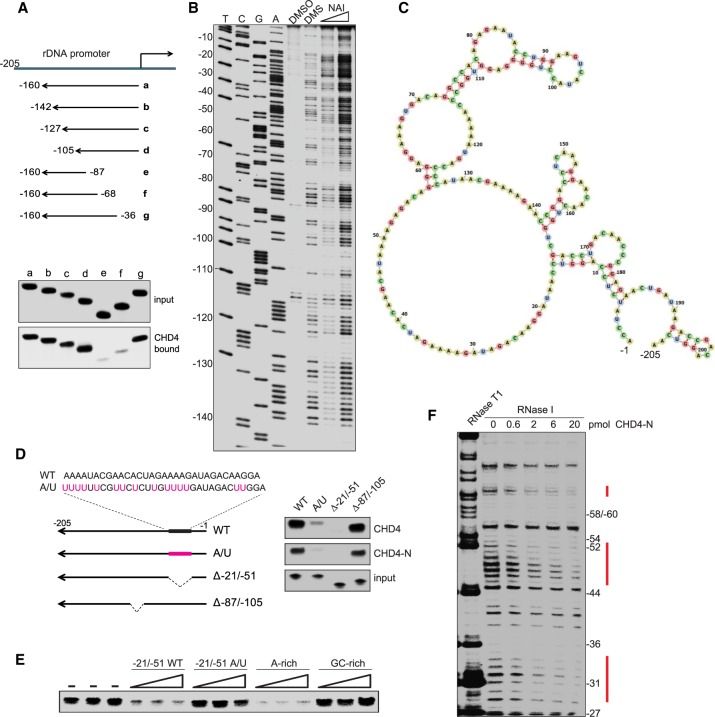
Figure 3.
CHD4 binds to A-rich single-stranded regions in PAPAS. (A) Pull-down assays comparing binding of truncated radiolabeled PAPAS to immobilized Fl-CHD4. (B) Secondary structure determination of PAPAS using dimethylsulfate (DMS) and SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) probing. Synthetic PAPAS treated with 1% DMS, 50 mM 2-methylnicotinic acid imidazolide (NAI), or DMSO was reverse-transcribed using a 5′ end-labeled primer (mrDNA −205/−185), and cDNA was analyzed on a 6% sequencing gel. Lanes T, C, G, and A represent sequencing ladders of mouse rDNA using the same 5′-labeled primer. (C) Secondary structure of PAPAS derived from SHAPE/DMS probing. (D) Pull-down experiment comparing binding of radiolabeled truncated or mutant versions of PAPAS with immobilized Fl-CHD4 or Fl-CHD4-N. (E) Pull-down/competition experiment. Increasing amounts (10-fold, 33-fold, and 100-fold molar excess) of synthetic RNA comprising sequences −21/−51 of wild-type PAPAS, mutant PAPAS (A/U), or an unrelated A-rich or G-rich RNA were included in pull-down assays to compete for binding of CHD4 to radiolabeled PAPAS (−1/−205). (F) RNase I footprinting assay showing binding of CHD4 to PAPAS. 5′-labeled PAPAS (−1/−205) was incubated with increasing amounts of Fl-CHD4-N and partially digested with RNase I, and the cleavage products were analyzed on a sequencing gel.