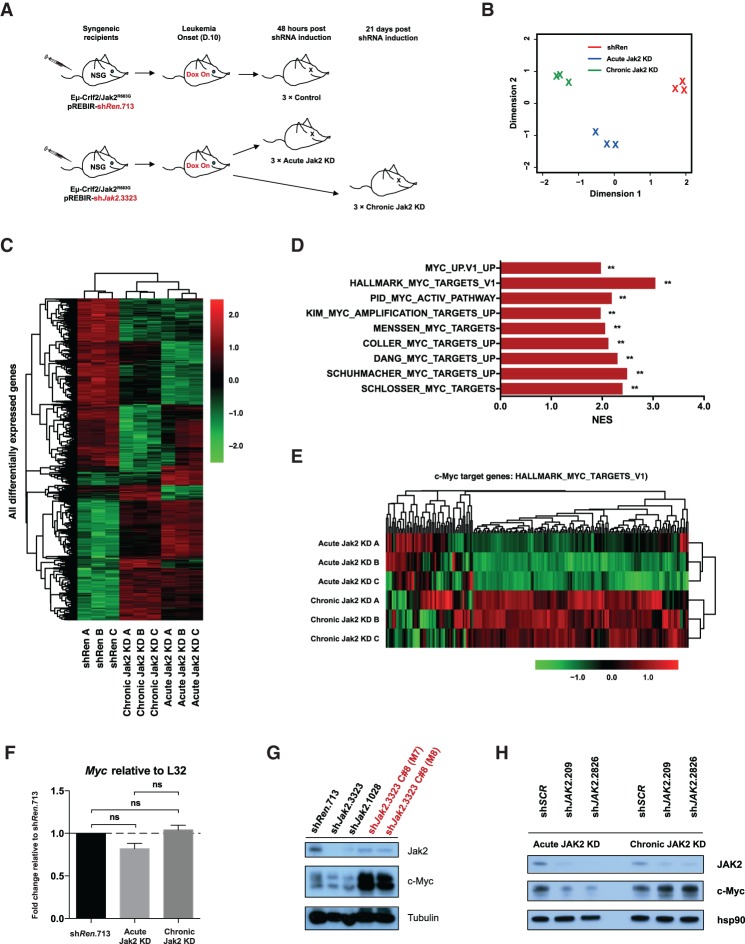
Figure 5.
Genome-wide transcriptome analysis of Jak2 knockdown-persistent Eμ-Crlf2/Jak2R683G-driven B-ALLs. (A) Schematic illustration of the generation of samples for RNA-seq. Dox-treated (day 10 after transplantation) recipient mice of Eµ-Crlf2/Jak2R683G-pREBIR-shRenilla.713 (n = 3) or Eµ-Crlf2/Jak2R683G–pREBIR-shJak2.3323 cells (n = 6) were sacrificed at either 48 h (acute knockdown) or 21 d (chronic knockdown) after shRNA induction, and RNA-seq was performed using RNA isolated from dsRed+/eBFP2+ splenocytes. (B) PCA (dimension 1 vs. dimension 2) of the top 500 most differentially expressed genes between naive (shRen; n = 3), acute Jak2 knockdown (n = 3), and chronic Jak2 knockdown (n = 3) Eµ-Crlf2/Jak2R683G B-ALLs. (C) Heat map of all significantly differentially expressed genes in all samples subjected to RNA-seq. Adjusted P < 0.05; log2 fold Δ ≥1 and −1 or less. The scale bar represents fold expression changes of each gene relative to the average gene expression across all samples. (D) Summary of normalized enrichment score values from GSEA performed on multiple independent established MYC gene signatures using genes differentially expressed between chronic and acute Jak2 knockdown Eμ-Crlf2/Jak2R683G B-ALLs. (**) FDR < 0.05. (E) Heat map of c-Myc target gene expression (HALLMARK_MYC_TARGETS_V1) in Eμ-Crlf2/Jak2R683G-driven B-ALL cells subjected to acute (48 h; acute Jak2 knockdown A–C) or chronic Jak2 knockdown in vivo (21 d; chronic Jak2 knockdown A–C). The scale bar represents fold expression changes of each gene relative to the average gene expression across all samples. (F) Quantitative PCR (qPCR) was performed using the RNA samples used for RNA-seq in A to determine relative mRNA levels of Myc in Eµ-Crlf2/Jak2R683G B-ALLs subjected to pREBIR-induced acute shRenilla.713, acute shJak2.3323 (acute Jak2 knockdown), or chronic shJak2.3323 (chronic Jak2 knockdown) expression in vivo. Error bars represent SEM. n = 2. (ns) Nonsignificant (P > 0.05). (G) Immunoblotting against the indicated targets was performed on lysates isolated from dsRed+/eBFP2+ splenocytes from mice transplanted with Eµ-Crlf2/Jak2R683G-pREBIR-shRenilla.713 (shRen.713), Eµ-Crlf2/Jak2R683G-pREBIR-shJak2.3323 (shJak2.3323), and Eµ-Crlf2/Jak2R683G-pREBIR-shJak2.1028 cells (shJak2.1028) at 48 h after shRNA induction and moribund bound Dox-treated tertiary recipients (shJak2.3323 C#8 [M7] and shJak2.3323 C#8 [M8]; red) of Jak2 knockdown-persistent cells (C#8) in Figure 4B. Tubulin served as a loading control. (H) Immunoblotting for the indicated targets was performed on lysates from GFP+ MHH-CALL4-pLMS-shSCR (shSCR), MHH-CALL4-pLMS-shJAK2.209 (shJAK2.209), and MHH-CALL4-pLMS-shJAK2.2826 (shJAK2.2826) cells at day 5 (acute JAK2 knockdown) and day 12 (chronic JAK2 knockdown) after transduction. Hsp90 served as a loading control.