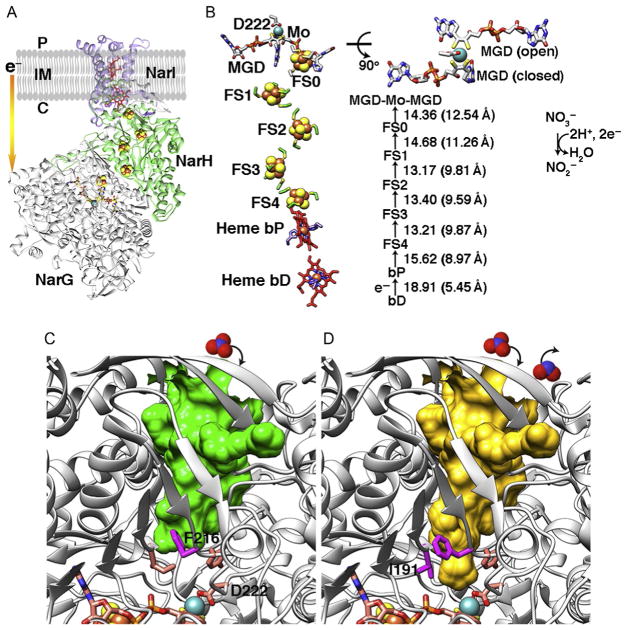
Fig. 6.
Cytosolic NarGHI structure from E. coli (PDB: 1q16) and comparative implications from PcrAB structures. (A) Overall structure of cNarGHI. The membrane-bound b-type heme protein NarI transfers electrons from the quinone pool to the cytosolic-facing nitrate reductase NarGH via the [Fe–S] clusters. (B) The layout and cofactor distances in cNarGHI are shown. MGD cofactor shows open and closed forms resembling those found in PcrA. (C) cNarG is in the closed state. Based upon our data and analyses, residue F216 is predicted to control the substrate tunnel access. (D) I191 may also be required to change conformation along with F216 to open full access to the active site. Substrate nitrate and product nitrite are shown in space-filling models (top).