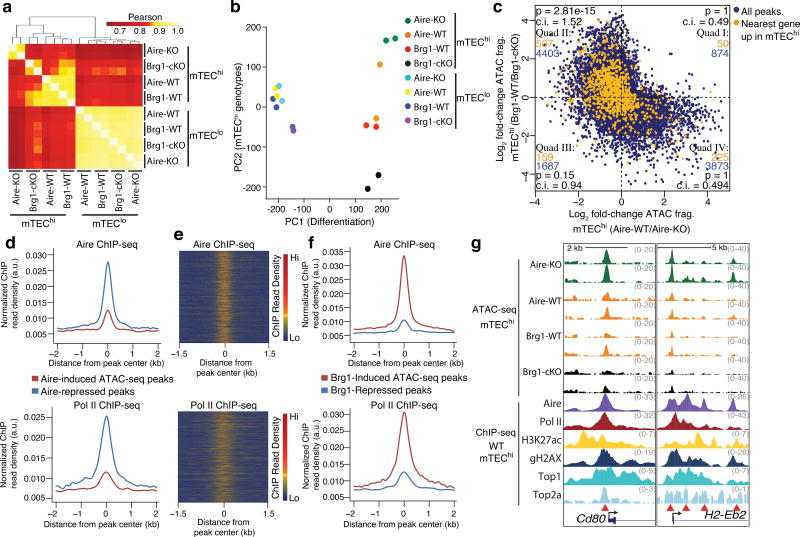
Figure 2.
Aire and Brg1 are determinants of an mTEChi-specific chromatin state with opposing influences on accessibility. (a) Correlations between indicated ATAC-seq samples at tissue-specific regions exhibiting induced accessibility during mTEChi differentiation. n = 4 independent experiments. (b) Principal component analysis of ATAC-seq data. (c) Comparison of the influences of Aire and Brg1 on mTEChi-specific accessibility state (ATAC-seq peaks correlated to PC2). Peaks whose nearest genes are upregulated in mTEChi vs. mTEClo are highlighted in orange and enumerated in each cartesian quadrant with total peaks (blue). P values indicate one-sided Fisher’s exact test with 95% confidence intervals (lower bound). (d) Aire (top) and Pol II (bottom) ChIP-seq fragment dyad density at Aire-induced or Aire-repressed ATAC-seq peaks. Representative of 2 independent experiments. (e) Heatmap of Aire (top) or Pol II (bottom) ChIP-seq fragment dyad density at Aire-repressed ATAC-seq peaks. Representative of 2 independent experiments. (f) Aire (top) and Pol II (bottom) ChIP-seq fragment dyad density at Brg1-induced or Brg1-repressed ATAC-seq peaks. Representative of 2 independent experiments. (g) Genomic signal tracks of ATAC-seq fragments at two loci from indicated mTEChi samples (top). ChIP-seq signal tracks (representative of 2 independent experiments) from WT mTEChi samples. Red arrowheads indicate differentially accessible regions.