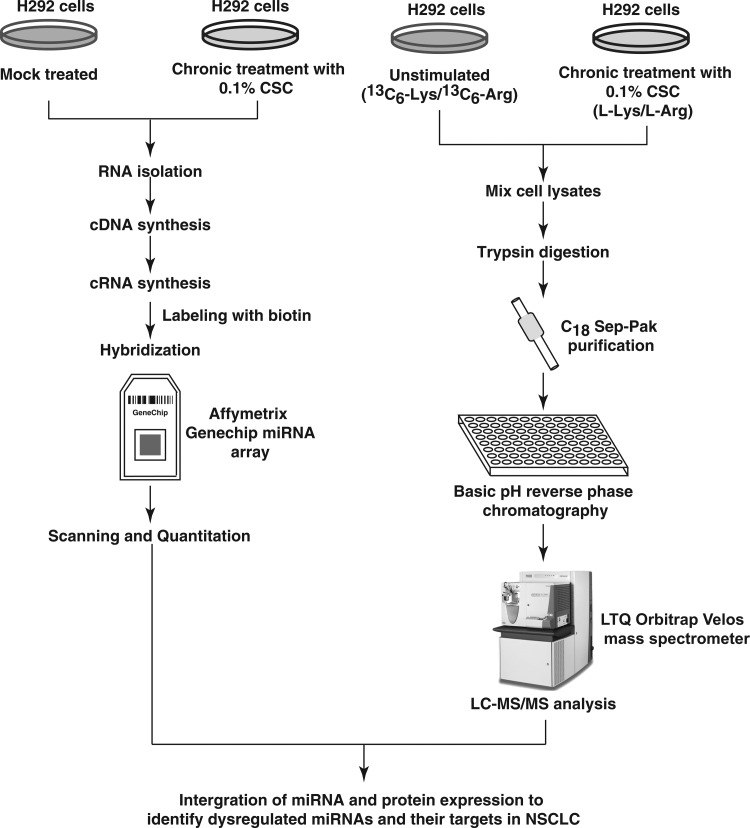
FIG. 1.
Experimental workflow employed to profile miRNA expression in smoke-exposed H292 cells. RNA was isolated from H292 cells, chronically treated with CSC. Samples were processed further depending on the integrity of the isolated RNA and labeling was carried out. The samples were hybridized on miRNA microarrays. Data were processed further to identify dysregulated miRNAs. Experimental workflow employed to analyze the proteome of smoke-exposed H292 cells. H292 cells were chronically treated with CSC. The cells were adapted to stable isotopic labeling with amino acids in cell culture media. The cells treated with CSC were grown in a medium containing normal arginine and lysine. The untreated cells were grown in a medium containing heavy arginine and lysine. Equal amounts of proteins from both treated and untreated samples were digested. The peptides were fractionated by strong cation exchange chromatography. The fractionated samples were subjected to LC-MS/MS analysis on a LTQ-Orbitrap Velos mass spectrometry. The data were searched and quantitated using the Sequest and Mascot search algorithms. CSC, cigarette smoke condensate; LC-MS/MS, liquid chromatography–tandem mass spectrometry.