Abstract
Advancing from statistical associations of complex traits with genetic markers to understanding the functional genetic variants that influence traits is often a complex process. Fine-mapping can select and prioritize genetic variants for further study, yet the multitude of analytical strategies and study designs makes it challenging to choose an optimal approach. We review the strengths and weakness of different fine-mapping approaches, emphasizing the main factors that affect performance. Topics include interpreting results from genome-wide association studies (GWAS), the role of linkage disequilibrium, statistical fine-mapping approaches, trans-ethnic studies, genomic annotation and data integration, and other analysis and design issues.
Subject categories: Biological sciences, Genetics, Genetic association study, Genome-wide association studies, [URI/631/208/205/2138]
Keywords: Biological sciences, Computational biology and bioinformatics, Statistical methods, [URI/631/114/2415]
Keywords: Biological sciences, Biological techniques, Genetic techniques, Genetic mapping, [URI/631/1647/1513/1382]
Keywords: Biological sciences, Genetics, Genetic linkage study, [URI/631/208/207]
Graphical Abstract
Fine-mapping is the process by which a trait-associated region from a genome-wide association study (GWAS) is analysed to identify the particular genetic variants that are likely to causally influence the examined trait. This Review discusses the diverse statistical approaches to fine mapping, their foundations, strengths and limitations, including integration of trans-ethnic human population data and functional annotations.
Introduction
Common complex human traits1 — including quantitative traits and diseases — often result from multiple environmental and genetic causes. Genome-wide association studies [G] (GWAS)1 have been widely used to identify the genomic regions on chromosomes that harbour genetic determinants of complex traits [G]2–8. To date, 47,681 statistical associations of single-nucleotide polymorphisms (SNPs) with complex traits are summarized in the US National Human Genome Research Institute (NHGRI)–European Bioinformatics Institute (EBI) GWAS Catalog9, representing 2,185 traits with association p-values less than 10−5. This success can be attributed to cost-efficient genotyping microarrays containing a large number of SNPs. However, the SNPs on microarrays typically do not cause10 the trait. Rather, the SNPs on microarrays, called tag-SNPs [G], are chosen because they are highly correlated (i.e., large amount of linkage disequilibrium [G] (LD)) with neighbouring SNPs, thereby serving as surrogates for large genomic regions that contain unmeasured SNPs11,12. The association between a tag-SNP and a trait can be indirect, resulting from tag-SNP associated with a causal SNP, which in turn is associated with a trait. Because patterns of LD among SNPs can be complex, it can be challenging to determine the underlying causal variants [G]. This is when fine-mapping [G] can help13. The principles discussed here are also relevant for common genetic variants measured by whole-genome sequencing studies.
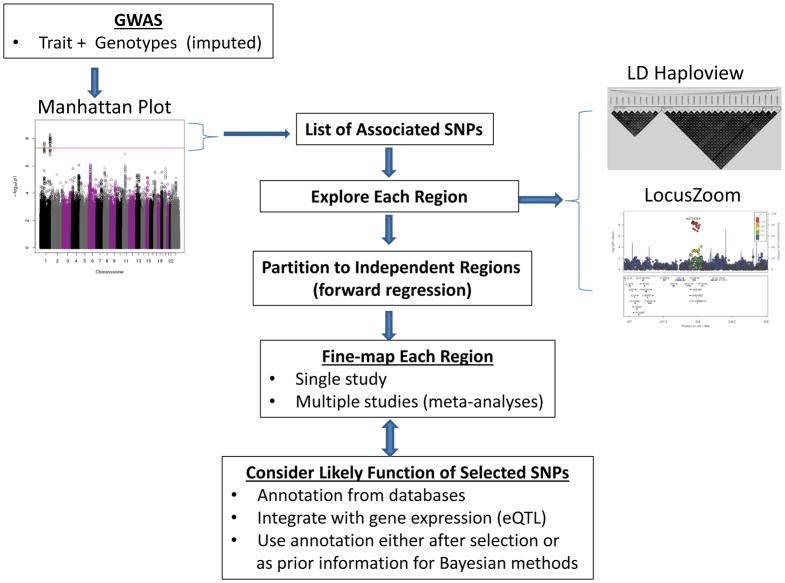
Fine-mapping seeks to determine the genetic variant (or variants) responsible for complex traits, given evidence of an association of a genomic region with a trait, and assuming at least one causal variant exists. After an initial GWAS identifies at least one SNP strongly associated with a trait (e.g., p-value <5×10−8), fine-mapping typically proceeds by the general steps outlined in Figure 1. These steps will be discussed in detail, but the general strategy is to use the GWAS list of SNPs associated with a trait to identify regions of interest. Each region is then visually explored for its LD structure and for genes known to be mapped to the region. Because it is simpler to fine-map one causal variant at a time, each region is partitioned into sub-regions that have approximately independent effects on the trait. Next, a fine-mapping strategy is chosen. The strategies we will discuss are illustrated in Figure 2. The selected SNPs are further evaluated for their likely function based on publicly available genomic annotation in order to prioritize costly and time-consuming laboratory-based functional studies.
Figure 1. Flow of typical process from intial GWAS to annotation of SNPs selected from fine-mapping analyses.
Based on GWAS p-values summarized in a Manhattan plot112, a list of SNPs that achieve genome-wide statistical significance (i.e., p-value <5×10−8) is used to determine regions of interest for fine-mapping. Each region is typlically explored according to the structure of linkage disequilibrium among single-nucleotide polymorphisms (SNPs) using Halpoview plots. Statistical assocaitions are viewed with LocusZoom plots that illustrate the patterns of association of each SNP with the lead SNP, as well as annotation of genes in the region. The regions can then be partitioned into independent sub-regions to ease computational burden, based on statistical models that evalute the simultaneous effects of multiple SNPs on a trait. Statistical fine-mapping is conducted in each region, using one of the methods illusrated in Figure 2. The SNPs selected from fine-mapping are then annotated with genomic features to prioritize follow-up functional studies. Figure is adapted from REF112.
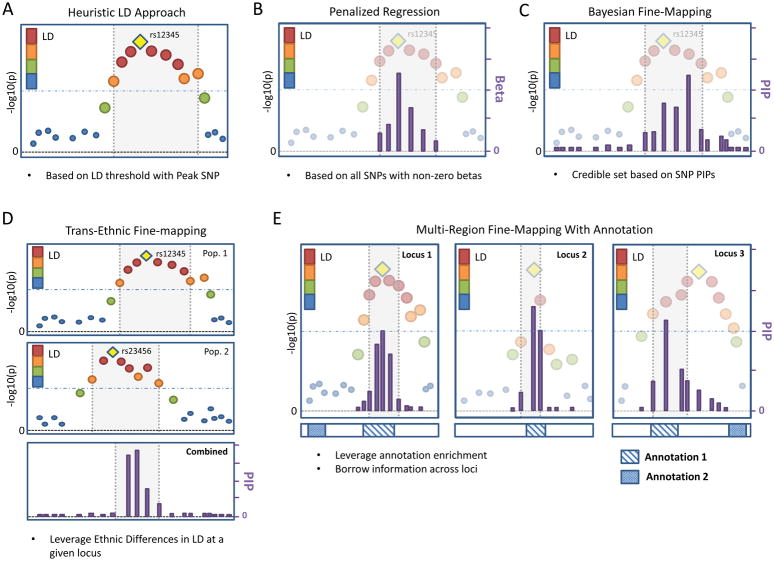
Figure 2. Hypothetical examples of fine-mapping strategies.
All subfigures are based on LocusZoom-style illustrations of marginal single-nucleotide polymorphism (SNP) associations. The −log10(p-values) are presented on the left y-axis and variant positions are on the x-axis. The gold diamond for each locus represents the peak SNP. The results for other SNPs are coloured by descending degree of linkage disequilibrium (LD) with the peak SNP (ordered red, orange, green, and blue dots). The purple bars represent additional variant-level statistics produced by fine-mapping (i.e., Beta values for penalized regression; posterior inclusion probabilities (PIPs) for Bayesian methods ), and the corresponding scale is on the right y-axis. The light grey boxes represent the regions selected by fine-mapping. A | The heuristic approach is based on LD patterns with the peak SNP (rs12345). All SNPs that meet the orange LD category threshold have been selected. B | The penalized regression approach selects all SNPs whose effects (Beta, right axis) are not shrunk to zero. C | Bayesian fine-mapping produces SNP-level PIPs (right axis), which can be summed to form credible sets based on a specified coverage probability threshold (e.g., 95%). This example illustrates that the peak SNP does not correspond to the SNP with the highest PIP, which can occur because of the correlation structure among all SNPs in the region. D | Bayesian trans-ethnic fine-mapping. Results for the same locus are illustrated for two diverse study populations (Pop. 1 (panel Da) and Pop. 2 (panel Db)) with different local LD structures. The peak SNPs for the two analyses differ (rs12345 versus rs23456), and combining the results through meta-analysis yields a narrowed fine-mapping credible region (panel Dc). E | Joint analysis of multiple loci by integrating annotation in Bayesian fine-mapping can improve fine-mapping by borrowing annotation information across loci. Presented are three example independent loci (panels Ea–c) along with two corresponding regional annotations, indicated by bands below each locus plot. For loci 1 and 2, the peak SNPs overlap with Annotation 1, indicating enrichment. This enrichment results in the SNP with the highest PIP in locus 3 to be different from the peak SNP in locus 3.
The main focus of this Review is on the statistical methods used to fine-map each region of interest. The overarching goal is to determine which variants are most likely to be functional, and to quantify the strength of evidence. This information can then be used for follow-up studies, such as large-scale replication studies of specific candidates, or laboratory functional studies. Although the initial GWAS can provide statistical evidence that a region is likely to harbour a causal variant, additional statistical methods are needed in order to discriminate likely functional variants from variants that are merely correlated with the functional variants. A variety of methods have been used, ranging from simple heuristic methods, to the use of penalized regression [G] for high-dimensional data, to more refined Bayesian methods tailored for fine-mapping (illustrated in Figure 2). Some of the methods can be applied to a single study, or to multiple studies combined by meta-analysis techniques. When combining multiple studies, special techniques have been developed to simplify sharing of data, based on summary statistics [G] 14. When the ethnic background of subjects differs across studies, trans-ethnic [G] fine-mapping can sometimes improve the resolution of fine-mapping. Each of these topics will be discussed, emphasizing their strengths, weaknesses, and challenges. We also present the main factors that influence power and resolution of fine-mapping, which in turn offer guidance for study designs. We review the use of genomic annotation for fine-mapping, as well as the integration of gene expression data with GWAS data. Finally, we discuss future challenges as our understanding of the genetic basis of complex traits evolves.
Interpreting lead SNPs from GWAS
The decision to fine-map a region typically follows the discovery of genome-wide significant results from a GWAS. It is common to summarize GWAS results with a Manhattan plot of all p-values that measure the marginal association of one SNP at a time with a trait, followed with LocusZoom plots for regions of interest15 (Figure 1). This allows one to focus on the SNPs with the smallest (that is, most significant) p-values in distinct regions, sometimes called the lead or index SNPs. GWAS results are most reliable when SNP associations achieve the accepted genome-wide statistical significance threshold of p-value<5×10−8 (REFS 16,17), a threshold that corrects for multiple testing, although some investigators use a weaker threshold of p-value<10−6 to highlight regions that are ‘suggestive’ of harbouring causal variants.
A limitation of the lead SNP is that there is a reasonable chance that it is not the causal variant. This can occur because GWAS microarrays are based on tag-SNPs, with the tag-SNP merely correlated with the unmeasured causal SNP. Furthermore, even if the causal SNP is measured or imputed, there is a reasonable chance that the statistical association of the causal SNP with the trait is not the most significant association among all the associated SNPs when statistical power [G] is not large. For example, based on simulations with 1,000 cases and 1,000 controls, there was a 79% chance that the lead SNP was found to be causal when the odds-ratio (OR) for disease risk was 1.5 and the risk allele frequency was 50%, but only a 2.4% chance when the OR was 1.1 and the risk allele frequency was 5%18. Zaykin et al.19 considered multiple causal variants and the impact of LD and drew similar conclusions that true associations are not likely to result in the smallest p-values, in part due to the small effect sizes of variants on complex traits. These findings emphasize the importance of caution when considering the lead SNP as likely causal, and the importance of fine-mapping in order to identify the causal variant(s).
LD for population fine-mapping
Fine-mapping in population-based studies leverages measures of non-random associations between pairs of loci. When loci are near each other and the frequency of recombination between them is low, the alleles from the different loci that occur on the same chromosome (called a haplotype [G] ) tend to be inherited as a unit. Alleles on a haplotype occurring together more than by chance is referred to as gametic association, or more commonly as linkage disequilibrium (LD). Various measures of LD have been proposed20, but they all depend on the difference between the observed joint frequency of alleles occurring on the same haplotype versus that expected by random chance. The most frequently used measure of LD is a standardized difference, which can be easily estimated by the Pearson correlation between the counts of the minor alleles (i.e., less common alleles) for two SNPs. This correlation coefficient is directly related to statistical power, and it is a reasonable measure for fine-mapping, although for case–control studies of rare diseases, measures such as attributable risk can perform better21.
Using LD to fine-map a complex trait is based on the premise that ancestral meiotic recombinations diminish LD, implying that the SNP with the strongest association with a trait is either the causal variant or close to the causal variant. However, evaluating one SNP at a time can be misleading due to the complex patterns of LD in a genomic region. A classic example is the association of Alzheimer’s disease with multiple SNPs around the APOE locus on chromosome 19. As the distance from APOE increases, the p-values for the association of SNPs with disease do not follow a monotonic pattern, but rather increase and decrease22. This opened a debate of whether there are genes in this region other than APOE that cause Alzheimer’s disease. Yet, the large amount of LD in this region has made it challenging to resolve this issue23. Factors beyond recombination that influence LD are mutation rates of the genetic markers, natural selection, population migrations and admixture, population bottlenecks, and demographic history (e.g., population size and mating patterns)24. Because LD is influenced by factors other than recombination, it is too limiting to rely solely on patterns of pairwise LD, or even haplotype blocks [G], to provide reliable fine-mapping of complex traits.
Factors influencing fine-mapping
A number of factors influence the performance of fine-mapping, including the number of causal SNPs in a region and their effect sizes on the trait, the local LD structure, sample size, SNP density, and whether the causal variants can be measured. Careful definition of the phenotype to enrich for genetic causes (e.g., disease severity, strength of family history) might increase the genetic effect size. The local LD structure is difficult to control, but trans-ethnic meta-analyses can capitalize on differences in LD structure. Factors that can be controlled in study designs are the sample size and SNP density. Sample size can be increased by pooling different studies or performing meta-analyses, recognizing that disease heterogeneity might increase, thus diluting efforts. Because it is critical to have high SNP density to capture the causal variants, in the following section we focus on strategies to increase density.
Increasing SNP density
The density of SNPs can be increased by DNA sequencing, but the costs for sequencing the large sample sizes required for fine-mapping can be prohibitive. Alternative cost-efficient strategies include genotype imputation [G] and additional genotyping.
Genotype imputation
Imputation of SNPs can fill in sporadic missing genotypes, harmonize data from different GWAS genotyping arrays to perform a pooled or meta-analysis, and increase the density of SNPs for fine-mapping25,26. Key criteria for imputation success are high correlation of directly assayed SNPs with the un-typed SNPs and appropriate reference panels that provide templates for LD patterns and allele frequencies that are representative of the study sample25. Popular reference panels are the 1000 Genomes Project27 and the Haplotype Reference Consortium28. Although imputed SNPs tend to be robust to the choice of quality control filters29, power to detect associations with a trait decreases as imputation accuracy decreases25.
Additional genotyping
Because imputation accuracy depends on the local LD structure, regions with weak LD might require actual genotyping to accurately evaluate their association with a trait. Situations where additional genotyping helps are: validation of imputed SNPs, possibly improving fine-mapping by reducing genotype measurement error; discovery of low-frequency SNPs that are not in strong LD with a lead SNP; identification of SNPs that are not well-represented in a reference panel; and disentanglement of SNPs that appear to be in perfect LD because of a small reference sample30.
Cost-efficient genotyping is becoming more accessible with the development of custom genotyping arrays that target certain diseases or traits, such as Oncoarray for common cancers31; Metabochip for metabolic, cardiovascular, and anthropometric traits32; and Immunochip for major autoimmune and inflammatory diseases33. These specialized arrays are primarily designed to evaluate associations of SNPs that are known to be associated with specific diseases or traits. Notable advantages of these arrays are increased density of SNPs in known gene regions and cost efficiency to achieve large sample sizes for fine-mapping. However, there are limitations. First, array content is focused on SNPs or genes that are already known to be associated with specific diseases or traits, which might exclude finding novel genes. These arrays also contain a backbone of genome-wide tag-SNPs that might overcome this limitation, albeit at a lower resolution than high-density arrays that are typically used for initial GWAS discovery. Second, array content can be based on reference data with incomplete coverage. Third, array content is based on SNPs rather than structural variation. Fourth, SNPs may be excluded from arrays due to assay failure. Finally, array designs are primarily based on European ancestry.
Partition to independent regions
When analyzing one SNP at a time, it is common to find multiple SNPs in a region to be marginally associated with a trait. This can occur when multiple non-causal SNPs are correlated with a single causal SNP. The LD among the SNPs will make each SNP appear to be associated with a trait when analyzing one SNP at a time. By contrast, when analyzing all SNPs jointly, only the causal SNP is expected to be associated with the trait when accounting for the LD among the SNPs, simplifying fine-mapping efforts. Alternatively, when there are in fact multiple causal SNPs, and they are correlated with each other and with other non-causal SNPs, fine-mapping can be more challenging because joint analyses are not expected to highlight just one causal SNP.
To simplify fine-mapping, it can be advantageous to first partition the region of interest according to the independent effects of SNPs on the trait, and then separately fine-map each partition. To determine if multiple marginally associated SNPs are driven by one or a few independent statistical associations, conditional forward stepwise regression is often used (e.g., logistic regression for case–control studies, or linear regression for quantitative traits). This entails conditioning on the lead SNP from a GWAS by treating it as an adjusting covariate in a regression model and testing the remaining SNPs in the region of interest. If a secondary SNP is found to be statistically significant after adjusting for the primary SNP, the sequential testing proceeds by treating some SNPs as adjusting covariates while screening the remaining SNPs, until no conditional tests are significant. A challenge is to decide on the threshold for significance. Some investigators use the stringent GWAS threshold p-value<5×10−8, while others use more liberal thresholds, such as p-value<10−4 or even p-value<0.05.
There are several limitations of forward stepwise conditional regression. First, as the number of steps increases, the number of statistical tests increases. If there are m SNPs, then after k sequential steps approximately km statistical tests will be performed, increasing the chance of a false positive result. Second, when m is large, perhaps close to the number of subjects in the sample, and when a liberal threshold is used to include SNPs at each step, the forward selection procedure becomes unstable34 and is overly optimistic regarding the trait variation explained by the selected SNPs35. Third, the probability (i.e., statistical power) to detect secondary signals diminishes as the correlation among SNPs increases.
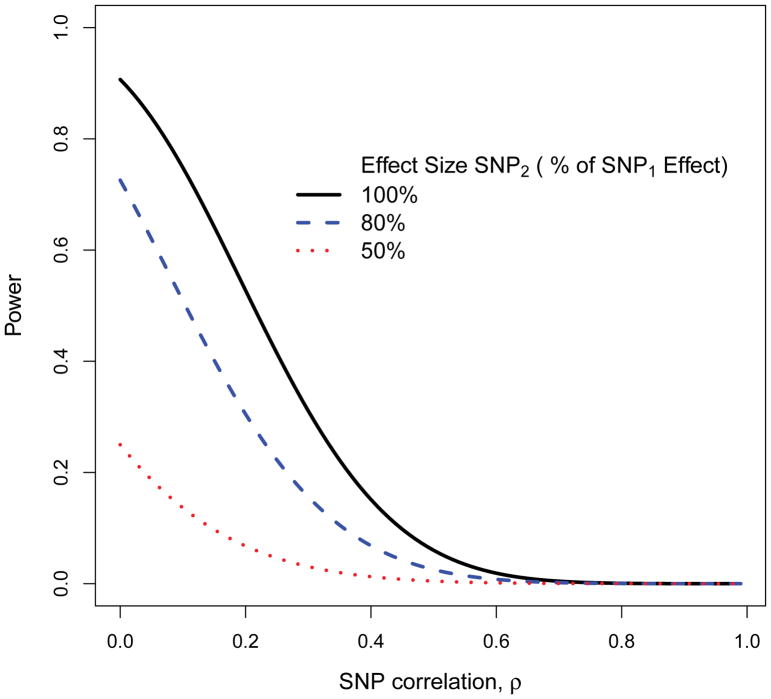
To illustrate diminishing power to detect a secondary SNP by conditional analysis, we derived simple formulas (see Supplementary information S1 (box)) to examine power. We assumed 90% power to detect the primary SNP when it explains 1% of a quantitative trait variation. We set the effect size for the secondary SNP to be a fraction of that for the primary SNP (fraction 50–100%), and we assumed the stringent GWAS threshold p-value<5×10−8 for testing the effect of the secondary SNP. Figure 3 shows a dramatic loss in power to detect secondary signals as the correlation of SNPs increases, even at levels of SNP correlation ρ=0.2. Power also decreases as the effect size of the secondary SNP decreases. Hence, one should be cautious when using conditional testing to declare only a single SNP to be statistically significant, because low power can cause missed secondary associations.
Figure 3. Power of conditional analysis.
This figure illustrates how conditional analyses have weaker power to detect secondary associated single-nucleotide polymorphisms (SNPs) compared to the power of an initial genome-wide association study (GWAS). Power of conditional analyses diminishes as the correlation of a primary SNP (indicated by SNP1) and a secondary SNP (indicated by SNP2) increases, and when the effect size of a secondary SNP is weaker than that for a primary SNP. For this figure, the power for an initial GWAS to detect a primary SNP1 is 90% for an effect size of R2 = 1% of explained trait variation. The effect size of a secondary SNP2 is varied from 100% to 50% of the effect size of primary SNP1.
Types of fine-mapping approaches
A number of approaches have been used to perform fine-mapping. We present three main strategies that have been used in the literature: heuristic methods, penalized regression models, and Bayesian methods (illustrated in Figure 2). Heuristic approaches were the first to be used, having grown out of practical experience and educated guesses, but with loosely defined criteria. Penalized regression models were developed in other fields of statistics, with the aim to reduce high-dimensional predictor variables (e.g., SNP data for fine-mapping) to a much smaller set that are strongly associated with a trait. In recent years, Bayesian methods have been specifically tailored for fine-mapping. The main features of these three approaches are discussed, along with their benefits and limitations.
Heuristic fine-mapping approaches
Because the LD structure around the lead SNP from GWAS has a substantial role in fine-mapping, it is popular to first examine the correlation among the SNPs surrounding a lead SNP. One approach is to filter SNPs according to their pairwise correlation (r2) with the lead SNP, retaining as potentially causal only those SNPs with an r2 above a threshold. Alternatively, hierarchical clustering of all SNPs in a region based on their pairwise r2 has been used to create clusters. However, both of these approaches depend on arbitrary thresholds to filter or form clusters. More rigorous penalized models and Bayesian methods that jointly model the simultaneous effects of multiple SNPs on a trait are more informative.
Another way to view correlation structure is by pairwise LD among SNPs within haplotypes, using software such as Haploview36 (Figure 1). This gives a visual impression of discrete haplotype blocks37. Combining the GWAS lead SNP with SNPs in the same haplotype block is an alternative way to select potential causal SNPs. However, one should be cautious with this approach. Recombination hot-spots [G] strongly influence block features, yet block boundaries can be arbitrary due to the choice of statistical model parameters and computational method, as well as genetic marker density and allele frequencies38,39. Internal inconsistencies of blocks are troubling, such as when two markers in strong LD flank markers with little LD with one of the flanking markers38. The above heuristic methods to choose SNPs for functional follow-up are too limiting because they do not account for the joint effects of the SNPs on the trait, and they do not give an objective measure of the confidence that a SNP is causal, but rather rely on somewhat arbitrary thresholds and subjective interpretations of correlations among SNPs.
Penalized regression models
An alternative fine-mapping approach is to use a regression model to jointly analyze all the SNPs in a region. Traditional model building is based on forward selection (or alternative stepwise methods), using p-values to determine whether a SNP should be included in a model. However, a large number of SNPs, and high correlation among the SNPs, can make traditional regression models unstable. A more robust approach is provided by penalized regression models. These models simultaneously perform estimation of SNP effect sizes and SNP selection into a model by shrinking small effect estimates toward zero. Popular penalized models are lasso40, elastic net41, minimax concave penalty42, and a normal-exponential-gamma shrinkage prior43 implemented in the hyperlasso software. Simulation studies show that penalized models tend to perform better than forward selection. Forward selection can be too conservative when using a very stringent p-value threshold to select SNPs, yet a liberal threshold increases the chance of falsely selecting SNPs44. Penalized models use tuning parameters to select SNPs into a model, with tuning parameters chosen to encourage SNPs with small effect sizes to be removed from the model. Tuning parameters are often estimated by cross-validation [G] to choose a model with minimum prediction error. Penalized models tend to result in sparse models, selecting only one or a few SNPs belonging to a group of correlated SNPs. This can result in a good prediction model that includes non-causal SNPs and excludes a causal SNP when they are highly correlated. In Supplementary information S1 (box) we provide R code for readers to further see how high correlation combined with sparse models reduces the chance of selecting the causal variant. By contrast, Bayesian methods have been specifically designed for fine-mapping, offering advantages over heuristic and penalized regression approaches.
Bayesian methods overview
The challenge of both penalized regression and Bayesian variable selection methods is to determine which SNPs have non-zero effect sizes (regression β values) on a trait. Although we refer to SNPs with non-zero effect sizes as causal, it is important to realize that statistical methods alone cannot determine causality. Penalized models choose SNPs based on cross-validation that minimizes the error of predicting a trait. By contrast, Bayesian inference focuses on the probability of a specific hypothesis, or specific model, thus providing probabilistic interpretation of models of interest.
A model for fine-mapping can be represented by an indicator variable for each SNP, with values of 1 for causal and 0 for not, and organizing these indicators for all SNPs of interest in vector c. For m SNPs, there are 2m possible c vectors (hence 2m possible models), ranging from all values of c equal to 0 for no SNPs causal, to all values equal to 1 for all SNPs causal. Using Bayes’ formula, the prior probability [G] of a model can be combined with the likelihood of the data D (trait and SNPs) to compute the posterior probability [G] of a specified model Mc, which we denote P(Mc | D). See box 1 for how the posterior probability is calculated. There are several ways to specify the prior probability for a model, such as assuming that variants are independent and equally likely to be causal45,46, or assuming a fixed number of causal variants out of the total variants46,47 (see Supplementary information S1 (box) for more details). The posterior probabilities for different models can be used to determine the posterior probability of including each SNP in any of the models (posterior inclusion probability [G] (PIP)), as well as determining the minimum set of SNPs required to capture the likely causal SNPs (credible sets). A variety of Bayesian fine-mapping methods have been developed, and are summarized in Table 1.
Box 1. Bayesian methods.
The goal of Bayesian methods is to compute the probability of a specific model, conditional on the observed data, denoted D, which includes the trait and SNPs. For a model Mc specified by a vector c of indicator variables, each indicator variable having values of 1 if a SNP is causal and 0 if a SNP is non-causal, the posterior probability of model Mc is:
where the sum in the denominator is over all models from a specified model space ℳ. For example, if the model space were to allow exactly one causal SNP out of k SNPs, there would be k possible c vectors, and k possible models in the space ℳ.
The term P(D|Mc) is the probability of the data under an assumed model. If only a fixed set of β values were considered, P(D|Mc) would be the likelihood of the data, denoted P(D|β). This is the same likelihood used in frequentist analyses. But, Bayesian inference treats the β values as random in order to account for their uncertainty. This is accomplished by assuming a prior distribution for the β values, which we denote P(β|Mc), and averaging over their possible values. This leads to the marginal likelihood, computed by integrating over the distribution of β values;
The ingredients of Bayesian methods are the likelihood [P(D|β)] and the assumed prior distribution of the models and their parameters. Details regarding choices of likelihood models and prior probabilities for Bayesian fine-mapping, as well as approximate Bayes factors, are discussed in Supplementary information S1 (box).
Table 1.
Commonly used Bayesian fine-mapping software.
| Software | Trait typea | Input covariatesb | Uses summary statistics? | Maximum number of causal variantsc | Input annotation? | Causal search | Main output | Refs |
|---|---|---|---|---|---|---|---|---|
| BIMBAM v1.0 | qt/binary | No | No | Fixed | No | Exhaustive | Bayes factor | 113;114 |
| mvBIMBAM v1.0.0 115,116 | mqt | No | Yes | 1 | No | Exhaustive | Bayes factor | |
| SNPTEST v2.5.4-beta3 | qt/binary/mqt/multinomial | No | No | 1 | No | Exhaustive | Bayes factor | 117 |
| piMASS v0.9 | qt/binary | No | No | Computed | No | MCMC | Bayes factor and PIP | 45 |
| BVS v4.12.197,118,119 | binary | Yes | No | Computed | Yes | MCMC | Bayes factor and PIP | 96,114,115 |
| FM-QTL | qt | No | No | Computed | Yes | MCMC | Bayes factor and PIP | 96 |
| DAP v1.0.0 | qt | Yes | Yes | 1/Fixed/Computed | Yes | Exhaustive | Bayes factor and PIP | 52 |
| Fine-mapping | multinomial | Yes | No | Computed | No | Greedy | PIP | 30 |
| Trinculo | multinomial | Yes | No | Computed | No | Greedy | Bayes factor and PIP | 30,120 |
| BayesFM | binary | Yes | No | 20 | No | MCMC | PIP | 30 |
| ABF | qt/binaryd | Yes | Yes | 1 | No | Exhaustive | Bayes factor | 121 |
| fgwas v0.3.6 | qt/binaryd | No | Yes | 1 | Yes | Exhaustive | Bayes factor and PIP | 95 |
| CAVIAR/eCAVIAR | qt/binaryd | No | Yes | Fixed | No | Exhaustive | ρ probability confidence set and PIP | 46,101 |
| PAINTOR v3.0 | qt/binaryd/mqt | No | Yes | Fixed/Computed | Yes | Exhaustive/MCMC | Bayes factor and PIP | 54,57,71 |
| CAVIARBF v0.2.1 | qt/binaryd | No | Yes | Fixed | Yes | Exhaustive | Bayes factor and PIP | 47,94 |
| FINEMAP v1.1 | qt/binaryd | No | Yes | Fixed | No | Shotgun stochastic search | Bayes factor and PIP | 53 |
| JAM in R2BGLiMS v0.1 | qt/binaryd | No | Yes | Fixed/Computed | No | Exhaustive/MCMC | Bayes factor and PIP | 55 |
Trait types are: binary, single binary trait; mqt, multiple quantitative traits; multinomial, trait with more than two categories; and qt, single quantitative trait.
For software that does not allow covariates to be input, the traits can be adjusted for covariates by first regressing out the covariates (i.e., subtracting trait predicted by covariates from trait values).
A fixed number is specified by the user, to reduce computational cost. It is usually small (e.g., ≤3) when the number of candidate variants is large. When computed, the number of causal variants is determined by the software. As indicated, some software allow different options for whether the maximum number of causal variants is fixed by the user or computed by the software.
Application to binary traits is based on linear regression, an approximation assuming small effect sizes and large sample sizes. MCMC, Markov chain Monte Carlo; PIP, posterior inclusion probability.
BIMBAM v1.0: http://www.haplotype.org/bimbam.html
mvBIMBAM v1.0.0: http://stephenslab.uchicago.edu/software.html#mvbimbam
SNPTEST v2.5.4-beta3: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
piMASS v0.9: http://www.haplotype.org/pimass.html
BVS v4.12.1: https://cran.r-project.org/web/packages/BVS
FM-QTL: https://github.com/xqwen/fmeqtl
DAP v1.0.0: https://github.com/xqwen/dap
Fine-mapping: https://github.com/hailianghuang/Fine-mapping
Trinculo: https://sourceforge.net/projects/trinculo/
BayesFM: https://sourceforge.net/projects/bayesfm-mcmc-v1-0/
fgwas v0.3.6: https://github.com/joepickrell/fgwas
CAVIAR/eCAVIAR: http://genetics.cs.ucla.edu/caviar/
PAINTOR v3.0: https://github.com/gkichaev/PAINTOR_V3.0
CAVIARBF v0.2.1: https://bitbucket.org/Wenan/caviarbf
FINEMAP v1.1: http://www.christianbenner.com/
JAM in R2BGLiMS v0.1: https://github.com/pjnewcombe/R2BGLiMS
Bayesian methods: posterior inclusion probability
Bayesian methods for fine-mapping have been specialized in order to focus on the SNPs that have the largest chance of being causal45,48. The PIP for a SNP is the probability of including a SNP as causal in any of the models. For SNP j, the PIP is computed by the sum of the posteriors over all models that include SNP j as causal,
After the posterior probabilities for the different models are computed, the PIP is rapid to compute and is an output from many Bayesian fine-mapping software packages (Table 1). Ranking SNPs by their PIP is a convenient way to select putative causal SNPs46. For example, the top k SNPs ranked by their PIP maximizes the expected number of causal SNPs across all possible subsets of size k47. However, caution is warranted when multiple SNPs in a region are highly correlated and all are approximately equally correlated with the phenotype. In this situation, none of the individual PIPs would be very large. It would be better to estimate the posterior expected number of causal SNPs by summing the estimated PIPs for all SNPs in the region45.
Bayesian methods: credible sets
Bayesian methods can be used to determine the α credible set49, the minimum set of SNPs that contains all causal SNPs with probability α. When assuming only one causal SNP, α is the sum of PIPs for SNPs in a set. This means that an α credible set is equivalent to ranking SNPs from largest to smallest PIPs50, and taking the cumulative sum of PIPs until it is at least α.
Bayesian methods can also allow for multiple causal SNPs in a region. Although computation time increases because of the larger number of models required for multiple causal SNPs, recent developments46,51–54, notably the JAM software55, make it feasible to stochastically search over a wide array of possible models. An excellent demonstration of the benefits of Bayesian fine-mapping is provided by a fine-mapping analysis of 84 prostate cancer GWAS loci using 143,804 subjects56. Single signals of causal variants were found in 63 regions, and 12 regions showed evidence of multiple independent causal variants. Furthermore, only 15 (5.4%) of the 280 original GWAS tag-SNPs remained as potential causal variants.
There are numerous advantages to Bayesian methods for fine-mapping. First, unlike p-values, posterior probabilities for SNPs can be directly compared. Second, they tend to select fewer SNPs as potentially causative compared to selecting SNPs based on their correlation with the lead SNP18. Third, simulation studies have shown Bayesian methods to perform better than both conditional stepwise regression46,55,57 and penalized regression models47. Finally, because Bayesian models are based on the joint effects of SNPs, they control for SNPs with large effects, improving power to detect SNPs with lesser effects. See Figure 2 for visual aspects of Bayesian fine-mapping.
Combining studies and meta-analyses
Combining data from multiple cohorts can increase fine-mapping resolution, and the fine-mapping strategies discussed above can be used when individual-level data are pooled. However, because it can be challenging to obtain individual-level data from multiple cohorts, summary statistics for the associations of a trait with SNPs can be used14. This strategy is gaining popularity because it simplifies data sharing and computational issues. When summary statistics are appropriately chosen, there is no loss of information compared to using individual-level data58. Most summary statistics focus on marginal regression β values and their variances. To obtain the joint effects of multiple SNPs using individual-level data, one must regress trait y on all SNPs simultaneously, y = XβJ+e, where βJ is a vector of the joint effects of SNPs, X is a matrix of the coded SNP effects (e.g., counts of minor alleles), and e is random error. By contrast, in summary data when only the marginal β values are available, it is still possible to determine the joint effects of SNPs needed for fine-mapping. When effect sizes are small, there is a simple relationship between the joint and marginal effects59, determined by βJ = R−1βM, where βM is a vector of the marginal effects and R−1 is the inverse of the matrix of pairwise SNP correlations (assuming the columns of X are standardized to have a mean of 0 and a variance of 1). The key aspect is the SNP correlation matrix, which summarizes the LD among the SNPs. When the original data are used to estimate the SNP correlations, there is no loss of information relative to analyzing individual-level data58. In practice, an appropriate reference sample is often used to estimate the SNP correlations, such as the 1000 Genomes Project, allowing marginal summary statistics from single SNP analyses to be combined for a joint analysis59. However, if the LD patterns in the reference sample do not represent those in the analyzed samples, the estimated joint effects can be biased, leading to errors in fine-mapping. Furthermore, the size of the reference sample should not be too small, and should increase in size as the GWAS sample size increases60.
Trans-ethnic fine-mapping
Comparisons of GWAS findings across ethnically diverse populations have revealed that associations of SNPs with complex traits are often consistent across populations, with similar direction of effects of alleles on traits61,62. Trans-ethnic meta-analyses that combine GWAS results of the same trait across genetically diverse populations can aid fine-mapping by capitalizing on ethnic differences in LD patterns63,64 (Figure 2).
A critical issue is the choice of ethnic groups. Analyses based on a mix of different European ancestries, or a mix of European and Asian ancestries, provide little gain for fine-mapping30,65. Rather, simulations have shown that a substantial reduction in the size of Bayesian fine-mapping credible sets can be attained by including subjects of African ancestry65, because they have much narrower LD. Including Hispanic ancestry might also improve fine-mapping over using only European or Asian ancestries65. The balance of ethnic groups can also impact fine-mapping performance. Simulations have shown that equal proportions of African and European ancestry is optimal for fine-mapping18. Although current samples with African ancestry tend to be much smaller than those with European ancestry, including African ancestry can still improve fine-mapping resolution65.
Trans-ethnic analyses are often conducted as meta-analyses of summary statistics using random-effects methods, recognizing that a SNP can have different effect sizes across different ethnic groups. This heterogeneity can result from differences in study designs, differences due to interactions with other SNPs, or differences due to environmental or lifestyle factors that influence the effects of genes. METASOFT66, which implements a modified form of the traditional random-effects approach, is popular among trans-ethnic studies67–70. Alternative methods that model the heterogeneity can improve fine-mapping resolution. One approach is based on Bayesian methods, as implemented in the PAINTOR software71. Other approaches that use GWAS SNP data to explicitly adjust for heterogeneity include MANTRA and MR-MEGA. MANTRA is based on a computationally intensive Bayesian partition method that creates discrete clusters of ethnically similar subjects, assuming that subjects within the same cluster have the same allelic effects for a variant, in contrast to subjects from different clusters having different allelic effects72. A good example of trans-ethnic fine-mapping with MANTRA, followed by use of PAINTOR and CAVIAR software to estimate PIPs and credible sets, as well as use of functional annotation, is provided in a large fine-mapping study of high density lipoprotein cholesterol73. In contrast to the discrete clusters created by MANTRA, MR-MEGA considers allelic heterogeneity on a continuum, and uses principal components of genetic similarity among subjects to determine axes of genetic variation, and then uses the principal components to adjust for heterogeneity of allelic effect sizes74. Simulations show that MR-MEGA can offer improved fine-mapping over other methods, yet retains computational efficiency74.
Factors affecting Bayesian PIPs for fine-mapping
A number of factors influence the resolution and power of fine-mapping. To quantify this influence, we derived a simple formula for the expected Bayes PIP of a single causal SNP when studying a quantitative trait (see Supplementary information S1 (box)). This expected posterior probability depends on the effect size of the causal SNP on a trait (measured by multiple regression R2, the percent of trait variation explained by the causal SNP) and the sample size (N). Importantly, these two factors combine into the non-centrality parameter that determines power, λ = NR2/(1−R2). Other factors that influence the expected posterior probability are the number of SNPs in the fine-mapping effort (we assume one causal SNP and m non-causal SNPs) and SNP correlation structure. To simplify the correlation structure, we assume that all SNPs are equally correlated with correlation ρ, as might occur when examining a small genomic region, or filtering on SNPs to achieve a correlation threshold. Finally, the assumed prior probabilities that SNPs are causal also influence the posterior probability. Based on these assumptions, the expected posterior probability for a causal SNP can be expressed as
| (1) |
Where pri is the prior probability that the ith SNP is causal, and subscript c is for the causal SNP.
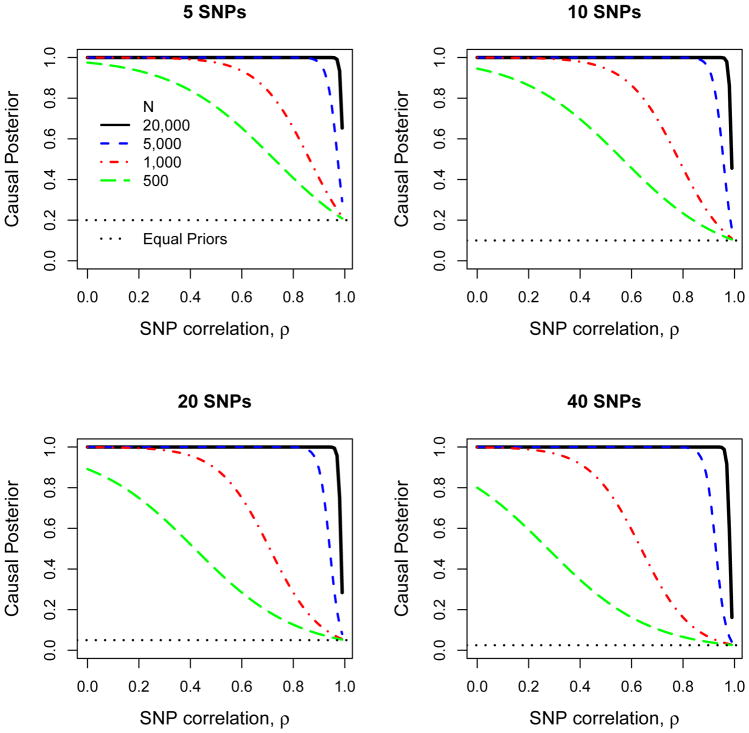
Assuming R2 = 1%, a relatively small effect size, we illustrate in Figure 4 that large sample size and small SNP correlations provide the ideal setting to achieve a high posterior probability for a causal SNP. Increasing the number of non-causal SNPs tends to decrease the posterior probability, by spreading out the posterior probability among multiple non-causal SNPs, particularly when the non-centrality parameter λ is not large. Figure 4 also emphasizes that it is difficult to achieve a large posterior probability for a causal SNP when SNPs are highly correlated. Hence, it might be premature to pre-filter SNPs to have a large correlation with a lead SNP before performing fine-mapping efforts, because the SNPs without large correlations could still be viable candidates. Furthermore, when power is weak or SNP correlation is large, well-informed prior probabilities are most critical because in these situations they have a larger effect on the posterior probabilities. Of course, mis-specified prior probabilities can result in a non-causal SNP having a large posterior probability, misleading fine-mapping efforts.
Figure 4. Posterior probability for a single causal SNP when 5–40 SNPs are in a region of interest.
The prior probability that a SNP is causal is assumed to be equal for all SNPs. Sample size (N) ranges from 500–20,000, and the percent of trait variation explained by the causal variant, R2, is 1%. SNPs are assumed to be equally correlated with magnitude ρ. The horizontal dotted line is for equal prior probabilities for SNPs, and the posterior probability approaches this line when the data have little information to distinguish causal from non-causal SNPs.
Genomic annotation
Genomic annotation that assigns biological function to DNA sequences can be informative about the likely function of SNPs selected by fine-mapping analyses, and can aid prioritization of follow-up functional studies. Large-scale initiatives have increased publicly available resources, including Gene Ontology75, GENCODE76, ENCODE77, FANTOM578, and the Roadmap Epigenomics Project79. By integrating multiple datatypes for a variety of tissues and cell-types, current annotations provide functional context for approximately 80% of the human genome80. Analyses of published GWAS findings have identified significant enrichment for functional annotations among complex trait associations81–83, motivating the use of annotation to improve fine-mapping resolution. Annotations are generally categorized according to protein-coding and non-protein-coding.
Protein-coding annotation
Annotation for SNPs in genes that code for proteins focuses on their impact on the resulting protein structure. Examples of annotation include whether a SNP occurs in an exon, an intron, a splice site, or whether it is involved in alternative splicing. A large number of bioinformatics annotation methods are available to functionally characterize coding SNPs and provide impact scores that predict their deleterious effect84,85. Although prediction accuracy can be low from individual methods, combining methods can lead to improved predictions, such as by the CADD86 and REVEL87 methods.
Non-protein-coding annotation
The ENCODE project has shown that the genome is pervasively transcribed, and that the majority of bases are found in primary transcripts, including non-protein-coding transcripts88. Genetic variation in non-coding regions is often involved in gene regulation. Some examples of non-coding annotation are promoters, enhancers, long non-coding RNA loci, transcription start sites, transcription factor binding sites, regulatory sequences, features of chromatin accessibility and histone modification patterns, and DNaseI hypersensitive sites. Variant impacts on putative transcription factor binding site (TFBS) motifs can be estimated by position weight matrices from databases such as TRANSFAC89 and JASPAR90. Annotation tools such as FIRE91, RegulomeDB92, and CADD86 provide regulatory impact scores for non-coding SNPs by aggregating multiple evidence sources of functional importance.
Integrating annotation into fine-mapping
Ad-hoc review of SNP annotations is often applied to SNPs selected by fine-mapping analyses in order to identify patterns of annotation enrichment and prioritize candidates for functional validation. This subjective approach can be cumbersome and biased. Alternative methods can improve fine-mapping, such as using functional annotation to weight SNPs93 in regression models, or extending Bayesian models to allow the prior probability that a SNP is causal to depend on annotation (see Supplementary information S1 (box))57,94–97. Advantages of Bayesian methods are that the weights given to functional annotation are inferred from the data, and jointly mapping multiple regions simultaneously provides a way to share annotation information across different regions57,95. Most methods allow for a relatively small number of annotation features, which requires screening each annotation one at a time in order to select a subset for a final model. An alternative approach that incorporates automatic selection of annotations from a large number of features overcomes this limitation94. Bayesian mapping software that allows use of annotation is highlighted in Table 1.
The broad impact of the use of annotation on fine-mapping is yet unknown. By simulations, Van de Brunt et al.18 found that incorporating annotation into Bayesian prior probabilities gave modest benefit for fine-mapping, by increasing the frequency of small credible sets (e.g., less than 10 SNPs) from 27% without annotation to 36% with annotation. In real applications of fine-mapping four blood lipid traits, the 90% credible set size reduced from an average of 17.5 SNPs without use of annotation to 13.5 SNPs with use of annotation57. When using a Bayesian method to fine-map three diseases in the Welcome Trust Case Control Consortium, very few SNPs in the credible sets were found to have annotated functions50. So, use of annotation information can be limiting from two angles: first, incorporation of annotation into prior probabilities has limited impact on well-powered studies and second, current understanding of broad genomic function may be too limiting to accurately improve prior probabilities of causation. Conversely, annotation might help when association signals are at best moderate, or in regions of high LD, when there are multiple causal SNPs in a region, or when different regions share enrichment for specific annotation features.
Integrating GWAS with gene expression
More than 90% of trait-associated alleles discovered by GWAS map to non-coding regions, with strong evidence of enrichment for regulatory elements, such as enhancers, promoters, insulators, and silencers81. Furthermore, SNPs associated with complex traits are significantly more likely to be expression quantitative trait loci [G] (eQTLs) than other SNPs on genotype arrays with the same allele frequencies98. This suggests that SNPs discovered by GWAS influence the amount of expression of nearby genes, and this altered expression ultimately influences the trait.
Statistical methods have been developed to integrate eQTL data with GWAS data to quantify the evidence of a causal pathway from SNP to gene-expression to a complex trait. The intermediate variable, mRNA, is the mediator between a SNP and a trait. One approach to test a causal pathway is by a causal inference test 99, with small p-values inferring causality. Alternative Bayesian approaches focus on the posterior probability that a single SNP is causally related to both mRNA level and the trait 100,101. Mendelian randomization is yet another approach that can be used to distinguish whether a single SNP influences both gene expression and the trait, versus whether separate SNPs in LD influence gene expression and the trait102. Details of these various approaches are provided in Supplementary information S1 (box).
A critical issue for integrating eQTL and GWAS results is the type of tissue for which the expression was measured. Complex diseases often result from dysfunction of multiple tissues or cell types, and the expression of genes varies widely across different types of tissues. Selecting relevant tissue types for a given disease process or complex trait can be a substantial challenge. Numerous public resources facilitate exploring gene expression in different tissues, such as the Genotype-Tissue Expression (GTEx) project, which includes genotypes, gene expression, and histological and clinical data for 449 human donors across 42 distinct tissues103.
Conclusions
Fine-mapping efforts have made considerable advancements to refine the most likely genetic variants discovered by large-scale genetic association studies of complex traits. We reviewed a variety of analytical methods, with the more sophisticated and relevant methods based on Bayesian fine-mapping. A common underlying basis of all methods is LD between measured SNPs and causal variants that makes fine-mapping feasible, as well as challenging. Although the details of a particular study will determine the success of fine-mapping, general guidelines for evaluating the robustness of fine-mapping efforts are provided in Box 2. Most of our Review discussed SNPs. Other types of genetic variants can be analyzed with similar strategies, such as small insertions or deletions (indels) analyzed as binary variables. Currently, there are few general methods to incorporate large structural variants into GWAS fine-mapping. Yet, germline structural rearrangements have helped to discover simple Mendelian disease genes, such as partial deletion of chromosome 15 leading to discovery of the gene UBE3A that causes Angelman syndrome104, or a 1.5Mb inversion on chromosome 17q12 leading to renal cysts and diabetes syndrome (RCAD), an autosomal dominant disorder 105. Genetic variants with more than two alleles would require extension of current methods. For example, the human leukocyte antigen (HLA) locus on chromosome 6 is highly polymorphic and is involved with immune response and multiple diseases. This region has unique fine-mapping challenges, some of which can capitalize on GWAS data (see Supplementary information S1 (box)).
Box 2. Guidelines for evaluating fine-mapping.
The goal of fine-mapping is to determine which variants in a genomic region are most likely to be causally related to a trait after accounting for how the variants in the region are correlated. One challenge with Bayesian fine-mapping can be that the output credible sets contain a large number of variants. A way to reduce the number of putative causal variants is improved use of annotation, perhaps by careful selection of relevant cell-types for gene regulation annotation. Another way is to increase sample size. With large samples, Bayesian fine-mapping can sometimes result in a credible set of just a few variants, or perhaps a single variant. To gain confidence in fine-mapping results, it is worthwhile to evaluate the robustness of the results. Some guidelines for evaluating robustness are given below.
What is the spread of the posterior inclusion probability (PIP) values for the variants in a credible set? If the PIPs are similar for multiple variants in a credible set, it can be worthwhile to view the correlation among the variants, and the effect sizes of the variants with effects of variants adjusted for each other. Large correlations among variants with similar effect sizes can suggest that it is not possible to distinguish among the variants with the available sample.
When using summary statistics and a reference panel to estimate the SNP correlations, how well does the reference panel match the study sample? When possible, it is worthwhile to compare the correlations among the variants in the study sample with the correlations in the reference panel. Large differences between study sample and reference panel warrant further fine-mapping analyses with the correlations obtained by the study sample. When correlations among SNPs from the study sample are not available, comparison of allele frequencies between reference panel and study sample can sometimes reveal problems, such as large differences is allele frequencies.
Because highly correlated SNPs can make fine-mapping difficult, sometimes compromising numerical computations, it is worthwhile to examine the correlations among the SNPs chosen based on fine-mapping. High correlations (e.g., more than 0.98) might warrant more careful scrutiny.
Different software for fine-mapping make different assumptions, and the computational methods can be implemented differently. To evaluate the robustness of a finding, it can sometimes be worthwhile to compare results across different software packages. Consistent results can improve confidence in the fine-mapping findings. However, inconsistent results can be caused by multiple factors, including different modelling assumptions, rounding errors, or differences in stochastic search methods. Hence, comparison of results across different software packages can be useful, but expertise in the software is required in order to give adequate interpretation of results.
Trans-ethnic fine-mapping assumes that the set of causal SNPs are the same for all populations, but accounts for differences in effect sizes across populations. For this reason, it can be worthwhile to evaluate the estimated effect sizes of the SNPs selected by fine-mapping to determine if just one, or a few, of several populations have effect sizes that dominate the fine-mapping.
As fine-mapping efforts proceed, an important consideration is the expected level of resolution. Using simulations that closely align with genomic structure of the UK 10,000 Genomes (UK10K) sample of 3,642 unrelated subjects of European ancestry, Wu et al.106 found that at least 80% of common variants identified in published GWAS that used imputed data were within 33.5 kb of causal variants. This provides optimism for fine-mapping GWAS results when using the study designs and computational methods discussed in this Review. Sharing of data in large meta-analyses, and improved genomic annotation, will probably improve resolution.
The frequency of variants has a large impact on the success of fine-mapping by statistical methods. Wu et al.106 found that rare untyped causal variants (minor allele frequency less than 0.01) are unlikely to map to a common variant in GWAS, whether using imputed data or whole-genome sequencing. Less-common variants tend to have lower levels of LD with other variants, making it challenging to impute less-common variants by statistical methods. But, low levels of LD can be advantageous for fine mapping when the variants are accurately genotyped or imputed and a study has high power to detect associations with single variants. Whole-exome sequencing and whole-genome sequencing open new avenues to discover rare variants. A challenge of these types of sequencing studies is the very large sample size (e.g., approximately 100,000 subjects) needed to discover regions that harbour rare causal variants for complex traits107. Furthermore, the statistical methods for rare-variant associations with a trait do not focus on single rare variants, but rather evaluate an entire region108. So, the initial discovery of a region provides little evidence of specific variants that are likely to be causal. Nonetheless, whole-genome sequencing can be informative for fine-mapping rare variants, but in this situation various sources of annotation of variants will be key to success109. Alternative approaches that combine functional assay results with computational prediction of function110 might prove useful for fine-mapping, albeit with the requirement of functional assays developed for each specific region of interest.
Our understanding of the genetic basis of human disease has evolved from single major genes influencing rare Mendelian disorders to multiple genes — polygenic — influencing common complex traits. Future fine-mapping challenges will face complex traits driven by a large number of variants with small effects, possibly based on a genetic architecture of regulatory networks composed of a small number of core genes that directly affect a trait, but with a large number of genes outside the core that indirectly affect the trait — an omnigenic model111. Understanding the genetic architecture of regulatory networks offers new opportunities to integrate this information into future fine-mapping strategies.
Supplementary Material
Online summary (Key Points).
Genome-wide association studies of complex traits can identify multiple associated loci, each represented by one or more single-nucleotide polymorphisms (SNPs) with small p-values. An important next step to understanding the genetic contributions to a trait is identifying which SNP(s) are likely to be truly causal at a given locus.
A major challenge in identifying underlying causal SNPs is the presence of linkage disequilibrium (LD), which can lead to highly correlated association results and multiple significant SNPs at a locus of interest. Fine-mapping methods aid this process by selecting and prioritizing variants most likely responsible for complex traits.
A variety of computational fine-mapping approaches are reviewed, including heuristic, penalized models, and Bayesian methods. Bayesian methods are highlighted because of their flexibility and benefits.
The main factors that influence the performance of Bayesian fine-mapping are quantified and discussed, including the number of causal variants in a region, the size of the effects of the variants on a trait, the SNP density, the local LD structure, and the sample size.
Fine-mapping can be improved by further data integration, such as combining summary statistics from multiple cohorts to increases sample size, trans-ethic studies that capitalize on differences in the structure of linkage disequilibrium, use of genomic annotation, and integrated analysis of gene expression data with genetic markers and traits.
Acknowledgments
This research was supported by the U.S. Public Health Service, National Institutes of Health, contract grants numbers GM065450.
Glossary
- Genome-wide association studies (GWAS)
Scans of genetic markers, typically single-nucleotide polymorphisms (SNPs), across DNA of many subjects to find variants statistically associated with a complex trait.
- Complex traits
Either quantitative traits (e.g., blood pressure, height) or common diseases (e.g., major cancers) that are caused by many genetic and environmental factors working together, each having a relatively small effect and few if any being absolutely required for disease to occur.
- Tag-SNPs
SNPs that are sufficiently correlated with neighbouring SNPs such that the tag-SNP serves as a surrogate for unmeasured SNPs.
- Linkage disequilibrium (LD)
Non-random association of alleles at different loci on a haplotype in a given population. LD is key to fine-mapping, because coinheritance without recombination of alleles from different variants implies that the variants are proximal on the same chromosome.
- Causal variants
Genetic variants that mechanistically contribute to diseases or quantitative traits, but are not fully penetrant in the sense that the variant may not be a sufficient cause in isolation.
- Fine-mapping
To refine the genomic localization of causal variants by the use of statistical, bioinformatic or functional methods.
- Penalized regression
A way to estimate regression coefficients by maximizing the log-likelihood of the data while placing a penalty that constrains the size of the regression coefficients, shrinking small coefficients toward zero, sometimes exactly to zero. Although this causes coefficient estimates to be biased, it improves the overall prediction of the model by decreasing the variance of the coefficient estimates.
- Summary statistics
Measures of statistical association between a trait and one or more single-nucleotide polymorphisms (SNPs) that summarize the size of effects of the SNPs on the trait, the variances of the effect sizes, and how the effect sizes are correlated among themselves. For case–control studies, summary statistics include the estimated log-odds ratios from logistic regression, the variances of the log-odds ratios, and the correlations among the log-odds ratios.
- Trans-ethnic
Genetic association study including subjects from more than one ethnic background.
- Statistical power
The probability of correctly rejecting a null hypothesis of no statistical association between a single-nucleotide polymorphisms (SNP) and a trait when in truth a statistical association exists. Power depends on the magnitude of the SNP effect, the sample size, and the p-value threshold for deciding statistical significance.
- Haplotype
A combination of alleles found on the same chromosome.
- Haplotype block
A set of highly associated alleles on a chromosome that tend to be inherited together.
- Genotype imputation
A method for estimating (‘imputing’) the unobserved genotypes of study subjects, both for individuals with missing or unreliable genotypes at a genotyped single-nucleotide polymorphism (SNP) and for all individuals at an ungenotyped SNP.
- Recombination hot-spots
Genomic regions where the rate of recombination is much higher than neutral expectation.
- Cross-validation
A technique to build a prediction model by randomly partitioning the sample into a training set to train the model (e.g., determine which single-nucleotide polymorphisms (SNPs) to include in a model), and a test set to measure its predictive performance (e.g., average squared prediction error). It is common to split the original sample into 10 equal size subsamples, use 9 to train and 1 to test, and repeat this process 10 times such that each of the 10 subsamples is used as a test sample, and then average the predictive performance over the 10 training subsamples.
- Prior probability
In Bayesian probability theory, the probability distribution assigned to parameters of interest, specified to represent prior knowledge of their values before observing the data.
- Posterior probability
In Bayesian probability theory, the updated probability distribution of parameters of interest, conditional on the observed data.
- Posterior inclusion probability (PIP)
The marginal probability that a single-nucleotide polymorphism (SNP) is included in any causal model, conditional on the observed data, thereby providing weight of evidence that a SNP should be included as potentially causative.
- Expression quantitative trait loci (eQTLs)
Genomic regions that harbour one or more nucleotide variants that influence the amount of expression of a gene.
- Type-I error
(Also known as false-positive rate). The probability of incorrectly rejecting the null hypothesis of no association between a SNP and a trait, thereby falsely concluding a true association.
- Multiple testing correction
When testing more than one statistical association, the probability of declaring at least one significant result increases as the number of statistical tests increases. If each of m independent statistical tests uses p-value<α to declare significance, then the chance that at least one of the m tests is found to be significant is approximately mα. Multiple testing correction maintains the overall chance of declaring at least one significant result by using more stringent p-value thresholds for each association tested. The Bonferroni correction uses p-value<α/m to test each association.
Biographies
Schaid Biosketch:
Daniel Schaid is the Curtis L. Carlson Professor of Genomics Research in the Division of Biomedical Statistics and Informatics at the Mayo Clinic in Rochester, Minnesota, USA. His research focuses on the development of statistical and computational methods for genetic analyses, and collaborations on genetic epidemology of complex diseases.
Daniel Schaid’s home page:
http://www.mayo.edu/research/labs/statistical-genetics-genetic-epidemiology
Chen Biosketch
Wenan Chen is a research scientist in the Department of Computational Biology at the St. Jude Children’s Hospital, Memphis, Tennessee, USA. His research focuses on the development of statistical and computational methods for genetic and genomic data analyses, especially those related to pediatric diseases.
Larson Biosketch
Nicholas Larson is an Assistant Professor of Biostatistics in the Division of Biomedical Statistics and Informatics at the Mayo Clinic in Rochester, Minnesota, USA. His research interests include statistical methods for genetic association studies, next-generation sequencing data analysis, and multi-omics data integration.
Footnotes
Competing Interests: None
Author contributions
All authors contributed to researching content for the article, discussing content and writing. D.J.S. was responsible for reviewing and editing the manuscript before submission.
References
- 1.Hardy J, Singleton A. Genomewide association studies and human disease. The New England journal of medicine. 2009;360:1759–68. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Olama AA, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature genetics. 2014;46:1103–9. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic acids research. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding K, Kullo IJ. Methods for the selection of tagging SNPs: a comparison of tagging efficiency and performance. European journal of human genetics : EJHG. 2007;15:228–36. doi: 10.1038/sj.ejhg.5201755. [DOI] [PubMed] [Google Scholar]
- 12.Stram D. Tag SNP selection for association studies. Genetic Epidemiol. 2004;27:365–374. doi: 10.1002/gepi.20028. [DOI] [PubMed] [Google Scholar]
- 13.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24:R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nature reviews. Genetics. 2017;18:117–127. doi: 10.1038/nrg.2016.142. This paper reviews the developments and progress of using summary statistics from genetic association studies to perform joint analyses of genetic variants, for use in fine-mapping, and to perform transcription-wide association studies (TWAS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England journal of medicine. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 17.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic epidemiology. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 18.van de Bunt M, Cortes A, Brown MA, Morris AP, McCarthy MI. Evaluating the performance of fine-mapping strategies at common variant GWAS loci. PLoS Genet. 2015;11:e1005535. doi: 10.1371/journal.pgen.1005535. Based on extensive simulations, this paper evaluated various factors that influence statistical fine-mapping, and provides guidance on design of fine-mapping studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaykin DV, Zhivotovsky LA. Ranks of genuine associations in whole-genome scans. Genetics. 2005;171:813–823. doi: 10.1534/genetics.105.044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin B, Risch N. A comparison of linkage diequilibrium measures for fine-scale mapping. Genomics. 1995;29:1–12. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 22.Martin ER, et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet. 2000;67:383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerreiro RJ, Hardy J. TOMM40 association with Alzheimer disease: tales of APOE and linkage disequilibrium. Archives of neurology. 2012;69:1243–4. doi: 10.1001/archneurol.2012.1935. [DOI] [PubMed] [Google Scholar]
- 24.Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nature reviews Genetics. 2008;9:477–85. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature Reviews Genetics. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Genomes Project C. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southam L, et al. The effect of genome-wide association scan quality control on imputation outcome for common variants. European journal of human genetics : EJHG. 2011;19:610–4. doi: 10.1038/ejhg.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. This paper applied three complementary Bayesian fine-mapping methods to a large data set and nicely illustrates novel methods and their interpretations, along with strategies for using annotation to interpret fine-mapping results. The supplemental material is particulary informative for computational strategies for Bayesian fine-mapping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amos CI, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS genetics. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nature reviews Genetics. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 34.Hocking R. A biometrics invited paper. The analysis and selection of variables in linear regression. Biometrics. 1976;32:1–49. [Google Scholar]
- 35.Freedman D. A note on screening regression equations. The Am Statistician. 1983;37:152–155. [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Daly M, Rioux J, Schaffner S, Hudson T, Lander E. High-resolution haplotype structure in the human genome. Nature Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 38.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nature Reviews Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz R, Halldorsson BV, Bafna V, Clark AG, Istrail S. Robustness of inference of haplotype block structure. J Comp Biology. 2003;10:13–19. doi: 10.1089/106652703763255642. [DOI] [PubMed] [Google Scholar]
- 40.Tibshirani R. Regression shrinkage and selection via the lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- 41.Cho S, Kim H, Oh S, Kim K, Park T. Elastic-net regularization approaches for genome-wide association studies of rheumatoid arthritis. BMC proceedings. 2009;3(Suppl 7):S25. doi: 10.1186/1753-6561-3-s7-s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breheny P, Huang J. Penalized methods for bi-level variable selection. Statistics and its interface. 2009;2:369–380. doi: 10.4310/sii.2009.v2.n3.a10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoggart CJ, Whittaker JC, De Iorio M, Balding DJ. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genetics. 2008;4:e1000130. doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayers KL, Cordell HJ. SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genet Epidemiol. 2010;34:879–91. doi: 10.1002/gepi.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, Stephens M. Bayesian Variable Selection Regression for Genome-wide Association Studies, and other Large-Scale Problems. The annals of applied statistics. 2011;5:1780–1815. This paper provides a Bayesian computational framework to consider a large number of causal variants. [Google Scholar]
- 46.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W, et al. Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics. Genetics. 2015;200:719–36. doi: 10.1534/genetics.115.176107. This paper links Bayesian fine mapping using summary statistics and using full data, and describes an efficient computational approach using only relevant variables for each candidate model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson MA, Iversen ES, Clyde MA, Schmidler SC, Schildkraut JM. Bayesian Model Search and Multilevel Inference for SNP Association Studies. The annals of applied statistics. 2010;4:1342–1364. doi: 10.1214/09-aoas322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlin B, Louis T. Bayesian Methods for Data Analysis. 3. Chapman and Hall/CRC; Boca Raton: 2008. [Google Scholar]
- 50.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nature genetics. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace C, et al. Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping. PLoS genetics. 2015;11:e1005272. doi: 10.1371/journal.pgen.1005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen X, Lee Y, Luca F, Pique-Regi R. Efficient Integrative Multi-SNP Association Analysis via Deterministic Approximation of Posteriors. American journal of human genetics. 2016;98:1114–1129. doi: 10.1016/j.ajhg.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kichaev G, et al. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017;33:248–255. doi: 10.1093/bioinformatics/btw615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newcombe PJ, Conti DV, Richardson S. JAM: A Scalable Bayesian Framework for Joint Analysis of Marginal SNP Effects. Genetic epidemiology. 2016;40:188–201. doi: 10.1002/gepi.21953. This paper builds on prior developments of Bayes methods for fine-mapping, and develops a computationally efficient method to explore a wide range of models that can include mulitple causal variants in regions of interest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dadaev T, Saunders E, Newcombe P, Anokian E, Leongamornlert D. Fine-mapping of Prostate Cancer Susceptibility Loci in a Large Meta-Analysis ldentifies Candidate Causal Variants. Nature Communications (accepted) 2017 doi: 10.1038/s41467-018-04109-8. This paper illustrates prartical approaches to fine-mapping many genomic regions using Bayesian methods, and illustrates the use of quantile regression to evalute how genomic annotation is associated with SNPs that have a large Bayes posterior probability of being cauasally related to prostate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS genetics. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. This is the first of a series papers regarding PAINTOR software for fine mapping, allowing multiple causal variants, summary statistics, and integrating functional annotations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genetic epidemiology. 2010;34:60–6. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics. 2012;44:369–75. S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benner C, et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. American journal of human genetics. 2017;101:539–551. doi: 10.1016/j.ajhg.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ntzani EE, Liberopoulos G, Manolio TA, Ioannidis JP. Consistency of genome-wide associations across major ancestral groups. Hum Genet. 2012;131:1057–71. doi: 10.1007/s00439-011-1124-4. [DOI] [PubMed] [Google Scholar]
- 62.Marigorta UM, Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013;9:e1003566. doi: 10.1371/journal.pgen.1003566. This paper illustrates that common genetic associations of complex traits are highly conserved across diverse ethnic populations and motivates the application of trans-ethnic analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li YR, Keating BJ. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 2014;6:91. doi: 10.1186/s13073-014-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaitlen N, Pasaniuc B, Gur T, Ziv E, Halperin E. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet. 2010;86:23–33. doi: 10.1016/j.ajhg.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asimit JL, Hatzikotoulas K, McCarthy M, Morris AP, Zeggini E. Trans-ethnic study design approaches for fine-mapping. European journal of human genetics : EJHG. 2016;24:1330–6. doi: 10.1038/ejhg.2016.1. This paper demonstrates that reductions in fine-mapping credible sets are heavily dependent on ancestral composition of contributing studies and emphasizes the importance of trans-ethnic study design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. American journal of human genetics. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Human molecular genetics. 2013;22:2303–11. doi: 10.1093/hmg/ddt064. [DOI] [PubMed] [Google Scholar]
- 68.van Rooij FJ, et al. Genome-wide Trans-ethnic Meta-analysis Identifies Seven Genetic Loci Influencing Erythrocyte Traits and a Role for RBPMS in Erythropoiesis. Am J Hum Genet. 2017;100:51–63. doi: 10.1016/j.ajhg.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franceschini N, et al. Variant Discovery and Fine Mapping of Genetic Loci Associated with Blood Pressure Traits in Hispanics and African Americans. PLoS One. 2016;11:e0164132. doi: 10.1371/journal.pone.0164132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larson NB, et al. Trans-Ethnic Meta-Analysis Identifies Common and Rare Variants Associated with Hepatocyte Growth Factor Levels in the Multi-Ethnic Study of Atherosclerosis (MESA) Annals of human genetics. 2015;79:264–74. doi: 10.1111/ahg.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kichaev G, Pasaniuc B. Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. American journal of human genetics. 2015;97:260–71. doi: 10.1016/j.ajhg.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–22. doi: 10.1002/gepi.20630. This paper introduces a Bayesian partition model framework for trans-ethnic fine-mapping by clustering study populations based on genetic simliarity in order to account for heterogeneity of allelic effects on a trait. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cannon ME, et al. Trans-ancestry Fine Mapping and Molecular Assays Identify Regulatory Variants at the ANGPTL8 HDL-C GWAS Locus. G3. 2017;7:3217–3227. doi: 10.1534/g3.117.300088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magi R, et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Human molecular genetics. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yon Rhee S, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nature Reviews Genetics. 2008;9:509. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 76.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 78.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159, 1161. doi: 10.1126/science.337.6099.1159. This paper leverages cell-line regulatory annotation to identify disease-relevant cell-types and reveals common genetic trait associations are enriched in functional DNA. [DOI] [PubMed] [Google Scholar]
- 81.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma M, et al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics. 2015;16(Suppl 8):S3. doi: 10.1186/1471-2164-16-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–30. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mudge JM, Harrow J. The state of play in higher eukaryote gene annotation. Nature reviews Genetics. 2016;17:758–772. doi: 10.1038/nrg.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eilbeck K, Quinlan A, Yandell M. Settling the score: variant prioritization and Mendelian disease. Nature reviews Genetics. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ioannidis NM, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. American journal of human genetics. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–41. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic acids research. 2014;42:D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ioannidis N, et al. FIRE: functional inference of genetic variants that regulate gene expression. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx534. Accepted, To Appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sveinbjornsson G, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nature Genetics. 2016;48:314–317. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 94.Chen W, McDonnell S, Thibodeau S, Tillmans L, Schaid D. Incorporating Functional Annotations for Fine-Mapping Causal Variants in a Bayesian Framework using Summary Statistics. Genetics. 2016;204:933–958. doi: 10.1534/genetics.116.188953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. American journal of human genetics. 2014;94:559–73. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wen X, Luca F, Pique-Regi R. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS genetics. 2015;11:e1005176. doi: 10.1371/journal.pgen.1005176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintana MA, et al. Incorporating prior biologic information for high-dimensional rare variant association studies. Human heredity. 2012;74:184–95. doi: 10.1159/000346021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS genetics. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC genetics. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS genetics. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hormozdiari F, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. American journal of human genetics. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu ZH, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics. 2016;48:481. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 103.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Magenis RE, Brown MG, Lacy DA, Budden S, LaFranchi S. Is Angelman syndrome an alternate result of del(15)(q11q13)? American journal of medical genetics. 1987;28:829–38. doi: 10.1002/ajmg.1320280407. [DOI] [PubMed] [Google Scholar]
- 105.Antonacci F, et al. Characterization of six human disease-associated inversion polymorphisms. Human molecular genetics. 2009;18:2555–66. doi: 10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu Y, Zheng Z, Visscher PM, Yang J. Quantifying the mapping precision of genome-wide association studies using whole-genome sequencing data. Genome biology. 2017;18:86. doi: 10.1186/s13059-017-1216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Auer PL, et al. Guidelines for Large-Scale Sequence-Based Complex Trait Association Studies: Lessons Learned from the NHLBI Exome Sequencing Project. American journal of human genetics. 2016;99:791–801. doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morrison AC, et al. Practical Approaches for Whole-Genome Sequence Analysis of Heart- and Blood-Related Traits. American journal of human genetics. 2017;100:205–215. doi: 10.1016/j.ajhg.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guidugli L, et al. Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. American journal of human genetics. 2018 doi: 10.1016/j.ajhg.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Haralambieva IH, et al. Genome-wide associations of CD46 and IFI44L genetic variants with neutralizing antibody response to measles vaccine. Human genetics. 2017;136:421–435. doi: 10.1007/s00439-017-1768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS genetics. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS genetics. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stephens M. A unified framework for association analysis with multiple related phenotypes. PloS one. 2013;8:e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shim H, et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PloS one. 2015;10:e0120758. doi: 10.1371/journal.pone.0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 118.Quintana MA, Conti DV. Integrative variable selection via Bayesian model uncertainty. Statistics in medicine. 2013 doi: 10.1002/sim.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quintana MA, Berstein JL, Thomas DC, Conti DV. Incorporating model uncertainty in detecting rare variants: the Bayesian risk index. Genetic epidemiology. 2011;35:638–49. doi: 10.1002/gepi.20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jostins L, McVean G. Trinculo: Bayesian and frequentist multinomial logistic regression for genome-wide association studies of multi-category phenotypes. Bioinformatics. 2016;32:1898–1900. doi: 10.1093/bioinformatics/btw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2008;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.