Figure 6.
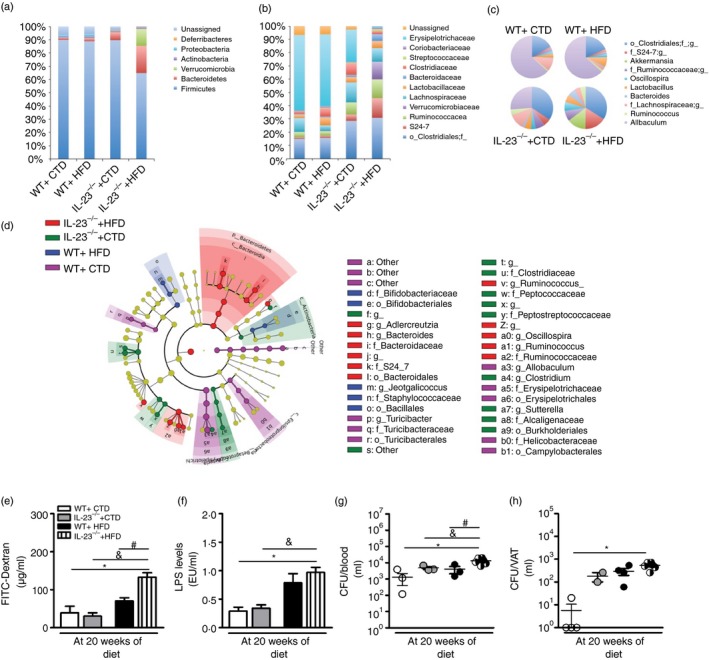
Gut dysbiosis, intestinal permeability and bacterial translocation in wild‐type (WT) and interleukin‐23 p19‐deficient (IL‐23p19–/–) mice on control diet (CTD) or high‐fat diet (HFD) for 20 weeks. Relative abundance of faecal bacterial phylum (a), family (b) and genus (c) were evaluated by 16S rRNA gene sequencing. Taxonomic cladogram comprising all detected taxa (d). Bacterial taxa that were significantly different among all pairwise comparisons were used as inputs for the Linear Discriminant Analysis Effect Size software. The rings of the cladogram stand for phylum (innermost), class, order, family and genus (outermost), respectively. Enlarged coloured circles are the differentially abundant taxa identified to be metagenomic biomarkers. Fluorescein isothiocyanate (FITC) ‐Dextran assay (e), serum lipopolysaccharide (LPS) levels (f) and colony‐forming unit (CFU) numbers in the blood (g) or visceral adipose tissue (VAT) (h). Asterisks represent statistically significant differences (*P < 0·05) compared with WT on CTD; (# P < 0·05) compared with WT on HFD; (& P < 0·05) compared with IL‐23p19–/– mice on CTD. Significant differences between the groups were compared by one‐way analysis of variance followed by Tukey's multiple‐comparison test. The results are representative of a single experiment repeated three times.
