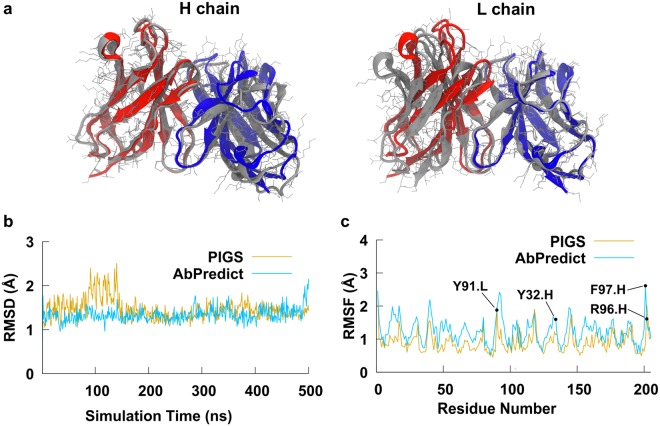
Figure 1.
Structural modeling of TKH2 monoclonal antibody. (a) Structure-based alignment of TKH2 mAb models with the heavy and light chains of PIGS model shown in red and blue respectively, and the AbPredict model shown in grey. The leftmost alignment is of the heavy chain (111 atom pairs, 0.8 Å RMSD), and the rightmost alignment is with the light chain (103 atom pairs, 0.7 Å RMSD). The overall RMSD of 1.57 Å indicates a difference in the heavy and light chain relative position between the two models. (b) An RMSD plot of both structure models Cα atoms relative to their initial structures over the 500 ns MD simulation. (c) Average atomic fluctuations of Cα atoms over the 500 ns simulations of PIGS and AbPredict structure models without the ligand. Residues that were selected to be flexible during one of the docking protocols are labelled (VH−Y32, VH−R96, VH−F97 and VL−W91).