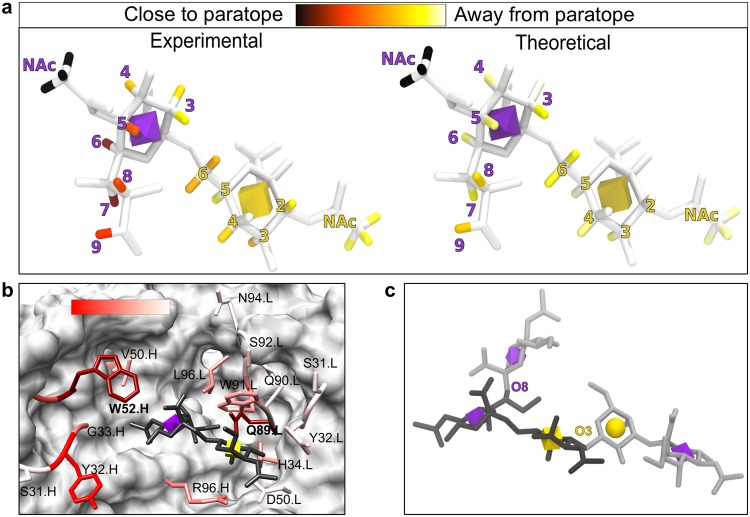
Figure 5.
Experimental and computational cross validation of ligand-Ab complex. (a) Graphical representation of normalized STD-NMR for STn antigen in complex with TKH2. The experimental STD-NMR values mapped onto a 3D structure of the antigen (Neu5Acα2–6GalNAcα-R). The antigen is shown in white licorice, with signal intensity for the hydrogens shown on a color scale (increasing signal intensity from yellow to red to black; atoms with no signal are colored white). A 3D-SNFG icon36 in the center of the residue ring depicts the Neu5Ac (purple octahedron) and Gal (yellow cube) residue. The figure was generated using VMD 1.9.351. (b) TKH2 binding site with residues experimentally mutated to alanine (licorice with a transparent surface). The residues are colored (white to red with a 2.2 gamma correction) by the change in apparent KD values (array data) for Neu5Acα2–6GalNAcα-ProNH2 binding when that residue was mutated to an alanine. Mutation of residues VL–Q89A and VH–W52A (burgundy) resulted in undetectable binding. The 3D structure shown is the final structure from the 100 ns MD simulation of the selected pose. (c) The two branch points (light grey licorice) from Neu5Acα2–6GalNAcα (dark grey licorice) found within the known human glycome are exemplified by glycan 9187 (glycomeDB ID). A glycan with extensions at either the Neu5Ac O8 or the GalNAc O3 are not predicted to fit in the TKH2 combining site based on the generated complex model. Glycans containing Neu5Acα2–6GalNAcα with extensions at the O4 of either the Neu5Ac or GalNAc have not been observed in humans.