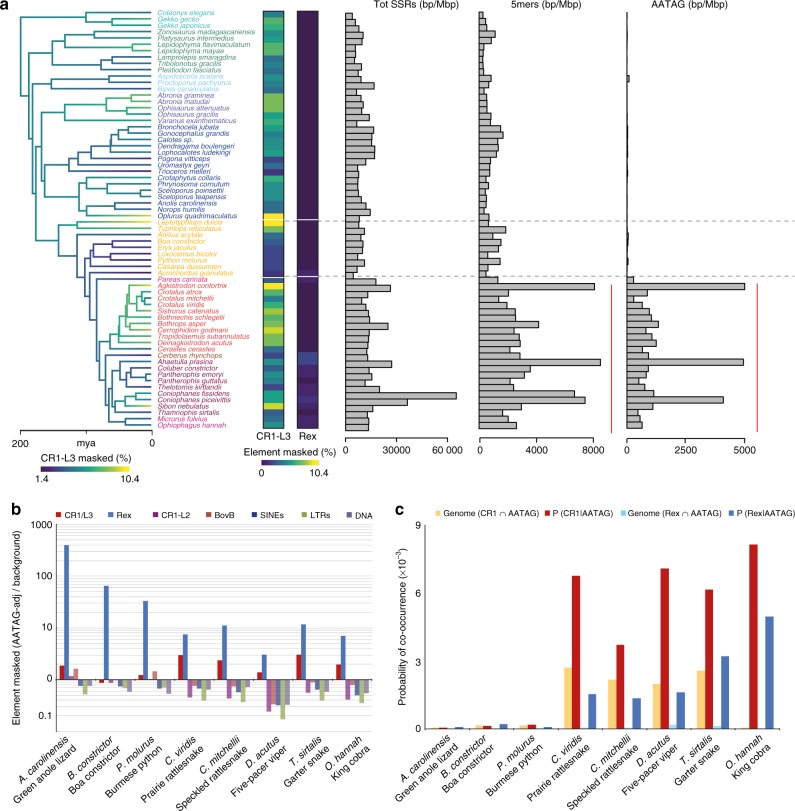
Fig. 2.
Microsatellite seeding by transposable elements (TEs) in squamate reptiles. a Branches on the time-calibrated consensus phylogeny are colored according to estimated rates of genomic CR1-L3 LINE evolution. Heat maps show the total genomic content (%) of LINE retrotransposon types involved in microsatellite seeding. Associated bar plots represent the total (left), 5mer (middle), and AATAG (right) microsatellite bp/Mbp density frequencies for each genome sampled. Red lines to the right of the bar plots highlight pronounced seeding of 5mer and AATAG microsatellites in colubroid snakes. b The ratio between TE mapping at the 5′ tail of AATAG microsatellite loci (AATAG-adjacent) and TE content averaged over five independent, randomly simulated genomic backgrounds for each class of TEs (SINEs; CR1-L3, Rex, CR1-L2 and BovB LINEs; LTRs; and DNA transposons). Ratios are plotted on a log scale to highlight enriched elements flanking AATAG loci (ratio >1) in contrast to elements more abundant in the genomic background (ratio <1). c Histogram shows joint and conditional probabilities of associations between AATAG loci and CR1-L3 and Rex. Genomic joint probabilities are shown in orange and light blue for CR1-L3 and Rex, respectively. AATAG-adjacent conditional probabilities are shown in red and dark blue for CR1-L3 and Rex, respectively