FIGURE 9.
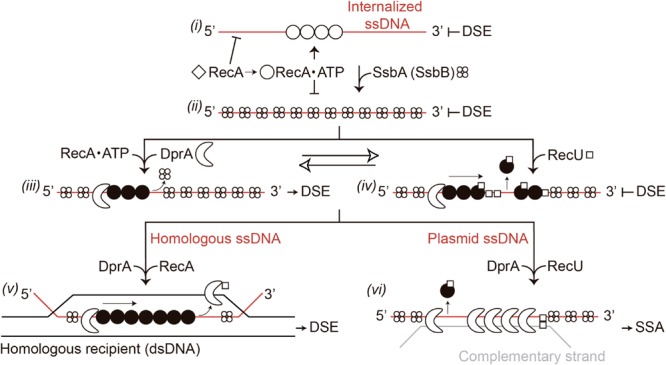
Model for RecA filament assembly on DprA-ssDNA-SsbA complexes in the presence of RecU. Apo RecA (empty diamonds) does not nucleate onto the internalized ssDNA. When ATP is present, RecA undergoes its first structural transition (empty circle). RecA-ATP can bind to incoming ssDNA and hydrolyze ATP, but cannot catalyze DNA strand exchange (DSE) (step i). When SsbA is present, RecA⋅ATP cannot nucleate on the SsbA-ssDNA complexes and cannot catalyze DNA strand exchange (step ii). Following interaction with DprA, SsbA recruits it onto the ssDNA. DprA in the DprA-ssDNA-SsbA complex interacts with and recruits RecA⋅ATP, to establish the 5′-end of the RecA filament. RecA undergoes its second structural transition (filled circle); and this active RecA assembles onto SsbA-coated ssDNA. Activated RecA remodels the SsbA-ssDNA complex to generate a DNA structure competent for SsbA displacement and RecA catalyzes DNA strand exchange in the presence of DprA-SsbA (step iii). RecU blocks RecA assembly on the ssDNA and promotes disassembly from the ssDNA (step iv). In the presence of homology with the recipient strand and a limiting concentration of DprA, it assists RecA⋅ATP assembly by displacing RecU to favor chromosomal transformation (step v). In the presence of plasmid ssDNA, DprA binds to plasmid ssDNA and, upon interaction with DprA-bound to a complementary incoming plasmid ssDNA (gray line), assists in the disassembly of RecA and RecU, and catalyzes single strand annealing (SSA) (step vi).
