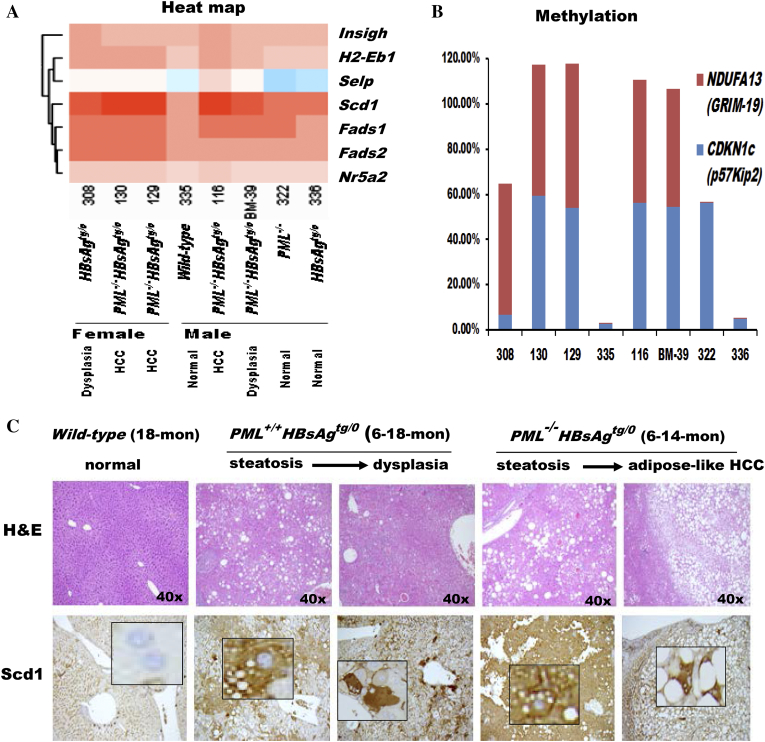
Figure 4.
Correlation of liver pathology with gene expression toward hepatocarcinogenesis in the livers of wild-type, HBsAgtg/0, PML−/− and PML−/-HBsAgtg/0 mice with different ages, sex and pathology. (A) Heat map demonstrating the cluster of differentially expressed genes related to lipid metabolism. Note that Scd1, Fads1 and Fads2 are desaturases involved in fatty acid synthesis and that Nr5a2 regulates cholesterol transport, bile acid homeostasis and steroidogenesis. (B) Epigenetic methylation analysis represented by percentage of growth control genes in livers from the same mice in (A). Note that, in addition to enhanced adipogenesis, PML loss is associated with impairment of the cell cycle and mitochondrial function during HBsAg-induced hepatocarcinogenesis. NDUFA13 (also called GRIM-19), a subunit of the mitochondrial NADH dehydrogenase, functions in the transfer of electrons from NADH to the respiratory chain and as a tumor suppressor by binding to STAT3. CDKN1c (also called p57Kip2) encodes a strong inhibitor of G1 cyclin/Cdk complexes and a negative regulator of cell proliferation. (C) Correlation of liver histology with immunohistochemistry of Scd1 protein expression in HBsAg-transgenic mice with or without PML loss. Note that PML loss induces severe steatosis with diffusely enhanced Scd1 expression and early-onset adipose-like HCC in HBsAg-transgenic mice. Squares represent in-situ zoomed regions.