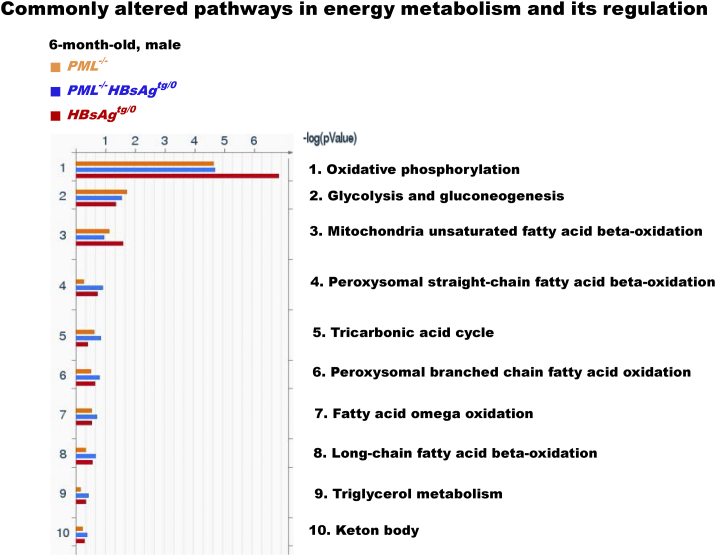
Figure S3.
The commonly altered metabolic pathways in the liver proteomic map folder of energy metabolism and its regulation. The enriched genes, whose expression was 2-fold up-regulated or down-regulated in more than 50% of the tested mice (n = 10 for each group) compared to the average expression of the same genes in sex- and age-matched wild-type mice (n = 5 for each group), were categorized by gene ontology (GO) pathway analysis. Note that oxidative phosphorylation is the most affected pathway in energy metabolism regulation.