 A DNA-programmed membrane fusion strategy for directing intracellular delivery of proteins into live cells.
A DNA-programmed membrane fusion strategy for directing intracellular delivery of proteins into live cells.
Abstract
Intracellular delivery of proteins provides a direct means to manipulate cell function and probe the intracellular environment. However, direct cytoplasmic delivery of proteins suffers from limited availability of efficient toolsets, and thus remains challenging in research and therapeutic applications. Natural biological cargo delivery processes, like SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex mediated membrane fusion and other vesicle fusion in live cells, enable targeted delivery with high efficiency. A surrogate of SNARE machinery represents a new direction in intracellular protein delivery. Here, we report a DNA-programmed membrane fusion strategy for guiding the efficient intracellular delivery of proteins into live cells. The inherent programmability of DNA hybridization provides spatiotemporal control of the fusion between protein-encapsulated liposomes and cell membranes, enabling rapid release of proteins directly into the cytoplasm, while still remaining functional due to the bypassing of the endosomal trap. We further demonstrate that delivered exogenous Cytochrome c effectively regulates the cell fate. Hence, this DNA-mediated fusion strategy holds great potential for protein drug delivery, regenerative medicine and gene editing.
Introduction
Introducing exogenous proteins into live cells plays an essential role in cell biological research and therapeutic applications.1 However, most proteins cannot directly diffuse through the lipid bilayer of cell membrane because of their hydrophilicity and large size.2 Although viral vector-based toolsets could achieve intracellular delivery of a DNA plasmid that encodes a protein of interest, this genetic approach lacks regulation and could induce stress responses, carcinogenesis, or immunogenicity.3,4 On the contrary, direct intracellular delivery of proteins represents a straightforward approach to modulate cell functions and treat diseases.1,5 For example, the plasmid expressed genome editing tool, the Cas9 protein, suffers from a high-frequency of off-target effects,6 and yet recent results suggest that intracellular delivery of functional Cas9 leads to higher genome modification and higher specificity compared to DNA transfection.7–9
However, unlike nucleic acid delivery, which is indirect and has been extensively studied, protein delivery suffers from a limited availability of efficient toolsets, and thus remains a challenge in research and therapeutic applications.10 Current intracellular cargo release tools, which are either physical membrane-disruption-mediated or carrier-mediated, are hampered by insufficient delivery efficiency and unpredictable cell function toxicity.8 Therefore, it is highly desirable to develop new intracellular delivery tools to target proteins for delivery into a wide variety of primary cells with integrated high efficiency and controllable cytotoxicity.
Membrane fusion is a universal process in live cells as it facilitates the transport of entities between and within cells.11,12 In live cells, membrane fusion is primarily mediated by SNARE complexes which facilitate lipid mixing, followed by pore formation and concomitant content transfer.13,14 Inspired by the SNARE machinery, several synthetic complexes have been developed as artificial membrane fusion systems.15–19 For example, Kros’s group has designed a fully synthetic membrane fusion system using a complementary pair of lipidated coiled peptides. The synthetic membrane fusion process delivers cargos directly into the cytoplasm, bypassing the endolysosomal pathway.20 They demonstrated the application of their artificial liposome-cell fusion system in a broad spectrum of small molecule delivery processes.21–25 Recent studies have revealed that DNA–lipid hybrids in a zipperlike hybridizing could also serve as artificial tethers to drag different liposomes into close proximity to drive membrane fusion.26 Here, we have demonstrated a DNA-programmed membrane fusion strategy for guiding protein delivery between live cell membranes and artificial liposomes on both suspension cells and adherent cells, and protein delivery was independent of the endolysosomal pathway. We also exemplified that cell fates could be regulated by DNA-mediated delivery of exogenous proteins.
Results and discussion
DNA-mediated fusion between liposomes and cell membranes and enzyme delivery
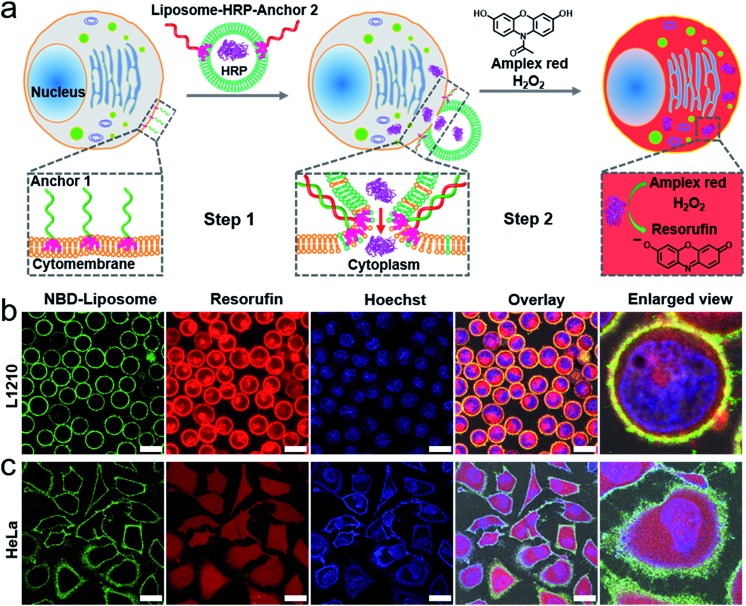
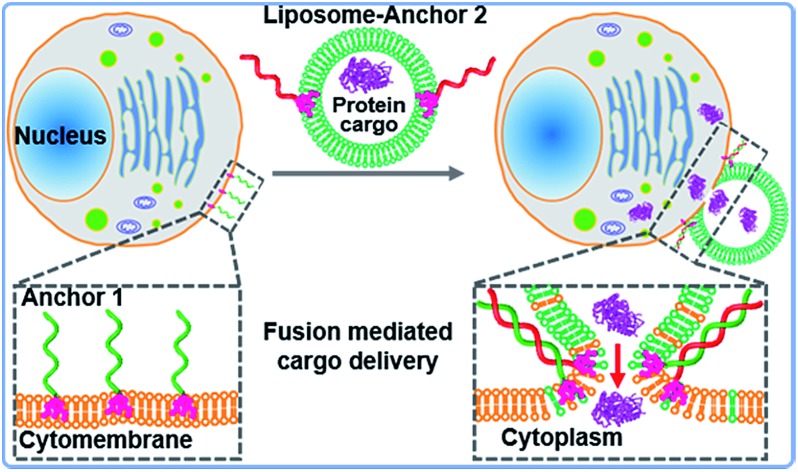
The principle of DNA-mediated membrane fusion is outlined in Fig. 1a. 3′cholesterol-functionalized single stranded (ss-) DNA (28 nt, anchor 1) and its complementary 5′cholesterol-functionalized ss-DNA (anchor 2) (Table S1†) were used to mediate fusion between live cell membranes and artificial liposomes (with a mean diameter of 100 nm, for the composition of the lipids, see Fig. S1 and the Experimental section†). In order to enhance the retention time of the DNA strands on the cell membranes,24 we compared the docking stability of the DNA variations with different cholesterol modification numbers, and found that ss-DNA with 3 cholesterol modifications had higher docking stability (Fig. S2 and S3†), thus ss-DNA with 3 cholesterol functionality at either 3′ or 5′ end was used in the present work to drag liposomes and cell membranes into close proximity. We found that cholesterol decorated ss-DNA could be easily inserted into cell membranes without perturbing the cell functions (Fig. S4 and S5†).27–29 Herein the orientation of cholesterol group was rationally designed at the same end of double stranded (ds-) DNA; thereby a zipperlike hybridization brings liposomes into close proximity to cell surfaces, which facilitates further membrane fusion.15,28
Fig. 1. (a) A schematic illustration of the DNA-mediated membrane fusion between the live cells and the liposomes, and the subsequent cytoplasmic delivery of active proteins. (b) and (c) confocal microscopy images of the L1210 and HeLa cells taken after the membrane fusion experiments. Green channel: the fluorescence of the NBD labelled liposomes, red channel: the fluorescence of resorufin (the product of HRP), blue channel: nuclear staining using Hoechst 33 258, overlay: the green, red and blue channels plus the bright field image and enlarged view: a single cell from the overlay channel. Scale bar: 15 μm in (b); 30 μm in (c).
In particular, we employed horseradish peroxidase (HRP, an enzyme extracted from plant cells) encapsulated within liposomes as an example of protein cargo (ca. 14 HRP per liposome, for detailed calculations see the Experimental section†). The motivation of using HRP as the protein cargo is its high enzymatic activity. Classical fluorophore cargos are less convincing for illustrating content mixing between small liposomes (<100 nm) and cells because fluorophore cargos in liposomes are strongly diluted in large cells, which makes them barely visible.30 HRP catalyzes the oxidation of a nonfluorescent substrate Amplex Red into a fluorescent product resorufin in the presence of H2O2. Amplex Red is an uncharged molecule that can passively diffuse across the lipid bilayer walls into the aqueous interior of cells, while the negatively charged fluorescent product resorufin cannot cross the phospholipid bilayer at neutral pH, which leads to its accumulation in the cellular interior.31 Moreover, H2O2 can also freely diffuse through cellular membranes. Therefore, upon delivering exogenous HRP into a cell, HRP could catalyze the fluorogenic reaction inside the cytoplasm, and in turn stain the cytoplasm.
We first performed a series of control experiments to confirm the solidity of this strategy. (1) We incubated Amplex Red or resorufin with untreated cells (L1210 and HeLa) in the presence of H2O2, and did not detect the fluorescence of resorufin (Fig. S6 and S7†). The lack of resorufin fluorescence confirmed that H2O2 did not increase the permeability of the cell membrane within the concentration range in our experiments (0.5 mM), and that intracellular HRP is the prerequisite for catalyzing the fluorogenic reaction. (2) We also incubated untreated cells (L1210 and HeLa) with free HRP (not encapsulated within liposomes), and then treated the cells with Amplex Red and H2O2. We found that the fluorescent products were clustered in the cytoplasm and remained in this cluster-like distribution for at least two hours (Fig. S8†). These results indicated that live cells could uptake HRP and that the fluorescent products are confined in the intracellular vesicles for a longer time rather than rapidly diffusing into the cytoplasm.
With the above results, we tested this strategy with both L1210 (suspension) and HeLa (adherent) cells (Fig. 1b and c), and found that liposomes incorporated with nitrobenzoxadiazole (NBD) fluorophore labelled lipids (NBD-PE) could efficiently dock onto the cytomembrane via zipperlike DNA hybridization and that the fluorescent products were evenly distributed inside not only the cytoplasm but also the nucleus of almost every cell (see amplified figure in Fig. S9†), which clearly suggested direct intracellular delivery of the protein cargos. Notably, there are many nucleopores on the nuclear membrane, therefore small molecules such as resorufin can get into the nucleus via passive diffusion. We noticed the red fluorescence of resorufin also located at the plasma membrane. According to a previous report on DNA-mediated fusion between liposomes and planar lipid bilayers, only 10% of the docked liposomes can fuse with the planar lipid bilayers within a short time,32 thereby the fluorescence of resorufin located at the plasma membrane was attributed to the liposomes that are still docked on the plasma membrane.
In addition, we emphasized that the zipperlike hybridization is the prerequisite for membrane fusion, while anti-zipperlike hybridization did not lead to efficient cargo release (Fig. S10†). Notably, there might also remain a possibility that HRP encapsulated within liposomes is released into the culture medium during the process of membrane fusion, and then diffuses into the cytoplasm via transient membrane destabilization during fusion.23 However, this possibility was excluded by the results of a control experiment incubating anchor 1 encoded cells with free HRP and empty liposomes carrying anchor 2, which indicated that the catalytic product resorufin was clustered in cells instead of evenly distributed (Fig. S11†). In another control experiment, in the absence of one or two of the anchor strands, the NBD-PE labelled liposomes could not be efficiently docked onto cell membranes, and the red fluorescence signal from resorufin was weak and clustered rather than being evenly distributed in the cytoplasm (Fig. S12 and S13†), indicating that liposomes containing HRP were taken up by cells via endocytosis and that the fluorescence products were confined in vesicles.
Protein delivery is a result of membrane fusion bypassing endocytosis
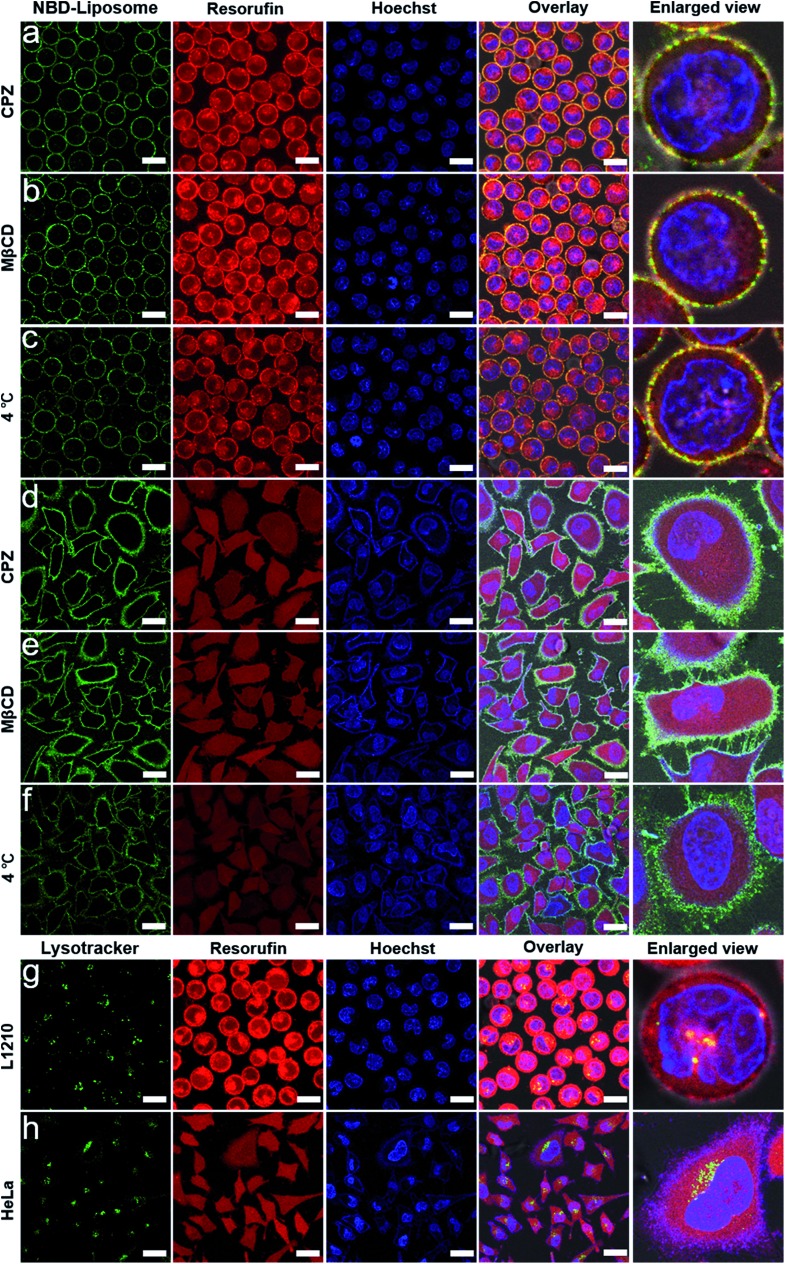
We next confirmed that the efficient intracellular protein delivery was a result of membrane fusion. Endocytosis is the primary pathway for cells to take in small particles including liposomes.10 Previous work has indicated that treating cells with endocytosis inhibitors could effectively inhibit various endocytotic pathways, for example, endocytic chlorpromazine (CPZ, interfering with clathrin-dependent endocytosis)33,34 and methyl-β-cyclodextrin (MβCD, disrupting caveola-dependent endocytosis).35,36 We also found that CPZ or methyl-β-cyclodextrin could reduce the cellular uptake of liposomes by L1210 and HeLa cells (Fig. S14†).
We thus performed membrane fusion experiments in the presence of CPZ or MβCD to investigate the possible endocytosis. However, we found that HRP encapsulated within DNA-encoded liposomes could still be efficiently released into cells, as confirmed by the evenly distributed red fluorescence of resofurin in the cytoplasm (Fig. 2a, b, d and e). In addition, control experiments indicated that neither CPZ nor MβCD treatment could change the permeability of the cell membranes which leads to the transmembrane diffusion of fluorescent resorufin and free HRP into the cytoplasm, even when the liposomes are docking onto the cell membranes (Fig. S15 and S16†). We next performed membrane fusion experiments at 4 °C based on the consideration that low temperature could shut down all possible endocytosis pathways. We found that HRP could be still delivered into cells, however, the delivery efficiency was lower, as indicated by the weakened fluorescence in cells (Fig. 2c and f), which was attributed to the decreased rate of membrane fusion at a low temperature.37 We also demonstrated that DNA hybridization did not improve the cellular uptake of liposomes (Fig. S17 and S18†), which excludes the possibility that improved cellular uptake of HRP loaded liposome results in an even distribution of fluorescent products inside cells.
Fig. 2. Confocal microscopy images of L1210 and HeLa cells taken after membrane fusion experiments but also treated with an endocytosis inhibitor or low temperature: (a) L1210/CPZ, (b) L1210/MβCD, (c) L1210/4 °C, (d) HeLa/CPZ, (e) HeLa/MβCD and (f) HeLa/4 °C. Green channel: the fluorescence of the NBD labelled liposomes, red channel: the fluorescence of resorufin, blue channel: nuclear staining using Hoechst 33 258, overlay: the green, red and blue channels plus the bright field image and enlarged view: a single cell from the overlay channel. Scale bar: 15 μm in (a–c); 30 μm in (d–f). (g) and (h) Confocal microscopy images of L1210 and HeLa cells taken after membrane fusion experiments and lysosome staining treatment. Green channel: the fluorescence of the lysotracker, red channel: the fluorescence of resorufin, blue channel: nuclear staining using Hoechst 33 258, overlay: the green, red and blue channels plus the bright field image and enlarged view: a single cell from the overlay channel. Scale bar: 15 μm in (g); 30 μm in (h).
We then stained the cells with lysotracker to localize the lysosomes in order to further exclude the possible endolysosomal pathway. We found that the lysosomes were scattered in the cytoplasm and that only a small fraction of resorufin could be colocalized with the lysosomes (Fig. 2g and h). These results further supported the conclusion that DNA-mediated fusion had dominantly released the protein cargos directly into the cytoplasm, bypassing the endolysosomal system.
Specific fusion of liposomes and different cells through specific DNA hybridization
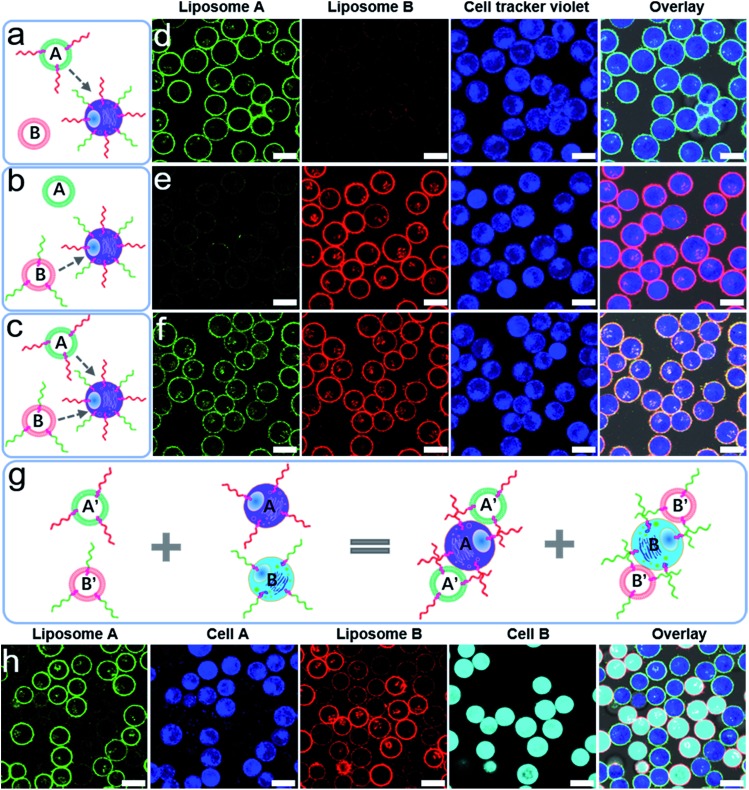
The richness of the DNA sequence and the specificity of the DNA hybridization suggests the possibility of directing the fusion between different DNA encoded cells and liposomes in a mixed system, which could be useful in intelligent protein delivery and cell function regulation.38–40 As depicted in Fig. 3, we designed a ternary system to address specific DNA-mediated fusion. In this ternary system, L1210 cells were decorated with two anchoring strands, anchor 1 and anchor 3 (3′cholesterol-functionalized DNA, see Table S1†), and stained by cell tracker violet. Two liposomes, NBD-PE labelled liposomes (liposome A, green) and Lissamine rhodamine (Rh)-PE labelled liposomes (liposome B, red), were encoded by anchor 2 (5′cholesterol-functionalized DNA which is complementary to anchor 1) and anchor 4 (5′cholesterol-functionalized DNA which is complementary to anchor 3), respectively. Only the presence of both anchor 2 and 4 resulted in the simultaneous docking of two liposomes onto the cell membranes (Fig. 3a–f).
Fig. 3. (a–c) A schematic illustration of the DNA encoded selective docking of liposomes onto L1210 cells in ternary systems. The red and green wavy lines represent cholesterol-functionalized DNA. (d–f) Confocal microscopy images of L1210 cells taken after mixing the liposomes and cells corresponding to the setup depicted in (a), (b) and (c). (g) A schematic illustration of the DNA encoded selective docking of liposomes onto L1210 cells in quaternary systems. (h) Confocal microscopy images of L1210 cells taken after mixing the liposomes and cells as depicted in (g). Green channel: the fluorescence of the NBD labelled liposomes, red channel: the fluorescence of Rh labelled liposomes, blue channel: cells stained by cell tracker violet BMQC dye, cyan channel: cells stained by cell tracker deep red dye, and overlay: the green, red and blue channels plus the bright field image in (d–f); the green, red, blue and cyan channels plus the bright field image in figure (h). Scale bar: 10 μm.
We further designed another quaternary system including L1210 cells by staining with cell tracker violet (cell A) and cell tracker deep red (cell B). The cells were decorated with anchor 1 (cell A) and anchor 3 (cell B). Two liposomes, liposomes A′ and B′, were encoded by anchor 2 and anchor 4, respectively (Fig. 3g). Upon mixing cells A and B with liposomes A′ and B′, we found that liposome A′ specifically docks onto cell A, while liposome B′ specifically docks onto cell B (Fig. 3h). Based on the above results, we further demonstrated specific docking and fusion in a mixed system, in which HRP containing NBD-PE labelled liposomes were encoded by anchor 2, while the cells were decorated with anchor 1 (cell A) and anchor 3 (cell B). We found that membrane fusion and enzyme delivery could only occur between anchor encoded liposomes and cell A (Fig. S19†). These results clearly highlighted the great potential of DNA hybridization for selective fusion in mixed systems.
Moreover, DNA hybridization-guided liposome docking greatly improved the efficiency of the cargo delivery. We compared the present strategy with a recently reported protein delivery strategy, carried out with the aid of cationic amphiphilic peptides. In the cationic amphiphilic peptide aided method, evenly distributed resorufin only appeared in a portion of the cells (Fig. S20†),41 which highlighted the efficiency of the DNA-mediated fusion strategy.
Programmable DNA hybridization enables programmable docking of liposomes onto cell membranes
Having confirmed that DNA could facilitate specific membrane fusion, we immediately realized that the inherent programmable nature of DNA hybridization42–52 provides versatile possibilities to mediate the programmable fusion and further programmable delivery of exogenous DNA or protein cargos into cells, which are urgently needed but are hardly available by other vectors. For example, one constraint on delivering protein drugs is a lack of delivery vehicles that allow for the controlled delivery of more than a single protein.53–55 Herein we have exemplified that the spatial and temporal fusion of liposomes with cell membranes could be rationally controlled by programmable DNA hybridization.
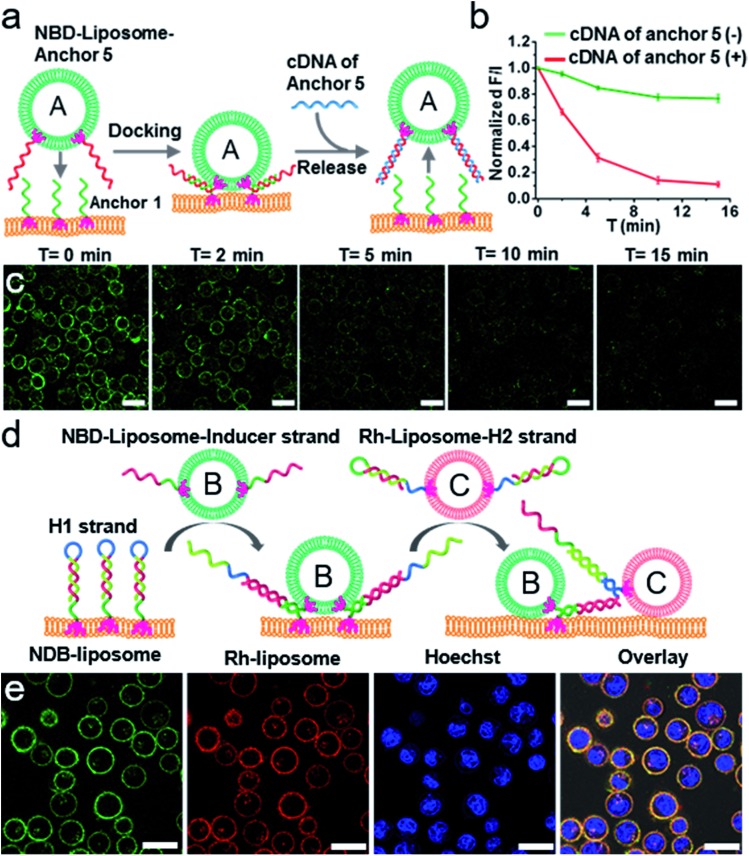
We first demonstrated the spatial control of membrane fusion by introducing a DNA strand (cDNA) that is fully complementary to anchor 5 which had encoded on the liposomes (Table S2†). Anchor 1 encoded on the cell surface is partially hybridized with anchor 5 (Fig. 4a). Thereby the spatial docking and fusion of the liposome encoded with anchor 5 could be relieved by a strand displacement reaction in the presence of cDNA. Indeed, upon the introduction of cDNA, the liposomes incorporated with NBD fluorophore labelled lipids were gradually released from the surface of the L1210 cells, leading to weakened NBD fluorescence on the cell surfaces (Fig. 4b and c, S21†). As described above, not all the docked liposomes would fuse with the cell membrane within a short time. However, the rate of the strand replacement reactions is generally rapid.56 Notably, in our study, the toehold length is 12 nt (Table S2†), therefore it is reasonable to rapidly remove the unfused liposomes from the cell membrane via the strand replacement reaction.
Fig. 4. (a) A schematic illustration of the spatial control of the docking and fusion of the liposomes with the cell surface by strand–displacement reactions. (b) The time-dependent normalized fluorescence intensity of NBD on the outside of the L1210 cells after the introduction of cDNA. (c) Confocal microscopy images showing the time-dependent fluorescence decrease of NBD on the outside of the L1210 cells upon the introduction of cDNA. Scale bar: 15 μm. (d) A schematic illustration of the temporal control of the sequential docking and fusion of two liposomes with cell surfaces by hybridization chain reactions. (e) Confocal microscopy images of L1210 cells. Green channel: the fluorescence of NBD labelled liposome B, red channel: the fluorescence of Rh labelled liposome C and the blue channel: nuclear staining using Hoechst 33 258. Overlay: the green, red and blue channels plus the bright field image. Scale bar: 15 μm.
In addition, we further demonstrated that the temporal control of the fusion could also be rationally controlled by the hybridization chain reaction, a strategy usually used to elongate DNA strands.57–59 As shown in Fig. 4d, the two liposomes B and C were incorporated with NBD and Rh fluorophore labelled lipids, respectively. Two hairpin strands (H1 and H2) were encoded on the surface of the L1210 cells and liposome C, respectively. An inducer strand was encoded on liposome B (Table S3†). By rational design, the hairpin structure has a short loop region and a long stem region which ensure a thermodynamic steady state. In the absence of inducer strands, the two hairpins fold into hairpin structures separately. Upon the introduction of the inducer strand, the steady state is disturbed by the hybridization of the inducer and H1, and thus the HCR reaction is triggered by the alternating sequential hybridization of H1, the inducer strands and H2. Therefore, upon mixing liposomes B and C with the L1210 cells, we observed the sequential docking of the liposomes onto the cell surface, as confirmed by the coexistence of NBD and Rh in Fig. 4e. Control experiments suggested that HCR is the prerequisite for inducing the programmable fusion of two liposomes on the cell surfaces; lacking either the inducer strands or the H2 strand did not result in the sequential docking of two liposomes onto the cell surfaces (Fig. S22†).
Intracellular delivery of exogenous protein through DNA-mediated membrane fusion to regulate cell fate
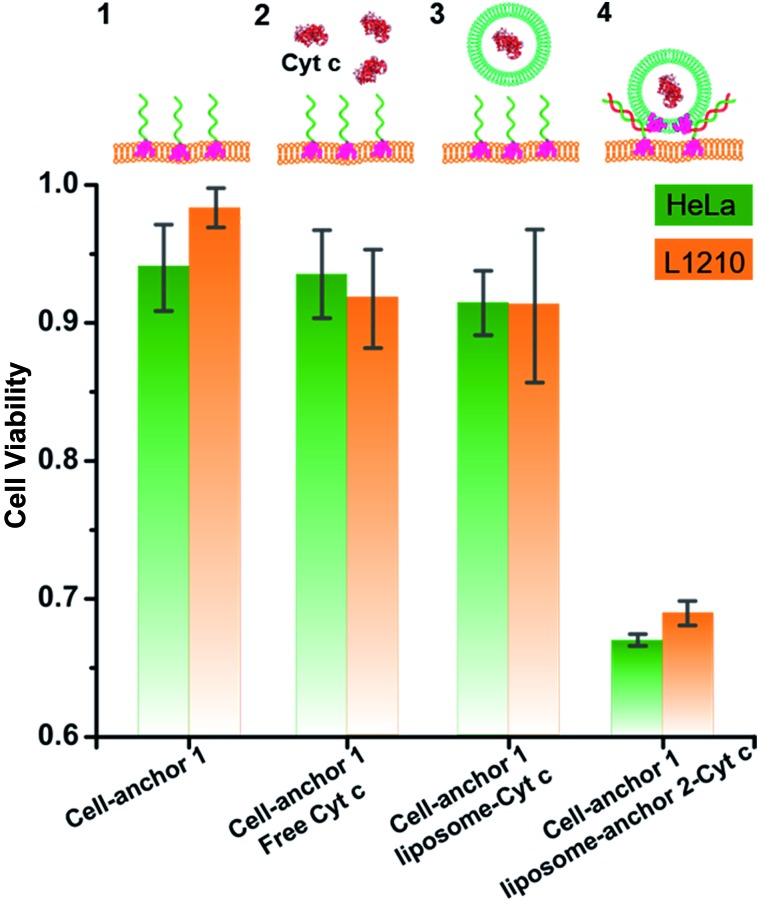
The ability to directly transfer protein into the cytosol suggests the possibility of regulating cell fate using exogenous proteins. Herein we exemplified this possibility with liposome encapsulated Cytochrome c (Cyt c). Cytochrome c is an important component of the oxidation respiratory chain. However, during the transduction of apoptotic signals, the alteration of the permeability of mitochondrial membranes causes the translocation of Cyt c into the cytoplasm, which in turn activates the death-driving proteolytic proteins, caspases.60,61 Therefore exogenous Cyt c is a model system used to challenge the cross membrane ability of various vector tools.62,63 We also delivered Cyt c into HeLa and L1210 cells using the present DNA-mediated membrane fusion strategy, and indeed found dramatically decreased cell viability (Fig. 5). Notably, the present fusion method bypasses the endosome–lysosome–lysosome escape pathway, which, in principle, enables the rapid delivery of protein cargos. In the present study, the 30 min of incubation time is much shorter compared with those in other endocytotic-dependent delivery vectors (see the Experimental section†),64,65 suggesting an advantage for the rapid delivery of protein drugs in therapy.
Fig. 5. The viability of L1210 and HeLa cells upon different treatments: (1) incubating the cells with anchor 1; (2) preincubating the cells with anchor 1, followed by treating the cells with free Cyt c; (3) preincubating the cells with anchor 1, followed by treating the cells with liposomes encapsulating Cyt c; (4) preincubating the cells with anchor 1, followed by treating the cells with anchor 2 encoded liposomes encapsulating Cyt c. Data are presented as the mean ± SD of 5 parallel experiments.
Conclusions
In conclusion, we have developed a biomimetic membrane fusion strategy via DNA hybridization, which enables targeted, programmable fusion between liposomes and cell membranes. The HRP based signal amplification strategy enables us to easily detect membrane fusion and direct intracellular delivery of protein cargos. The inhibitor treatment and lysosomal staining experiments suggested that the intracellular delivery of liposome encapsulated cargos bypassed the endocytotic pathway and went directly into the cytoplasm. This strategy could also be extended to delivering other bioactive macromolecular cargos, such as nucleic acids, protein/nucleic acid complexes, and small molecule drugs. In this work, we demonstrated only the in vitro applications, because it might be much harder to modify the target cells with cholesterol modified DNA in vivo. However, in vivo applications can be envisioned since DNA engineered cell surfaces have been extensively studied for enhancing the efficacy of cell therapy.66,67 For example, a previous study has indicated that lipophilic DNA could be inserted into the cytomembrane of tumor cells in vivo, and local injection of membrane anchored immunostimulatory oligonucleotides promoted the physical association of immunostimulatory oligonucleotides with tumor cells.68 This hints at potential in vivo applications of this strategy for DNA-programmed rapid delivery of exogenous proteins, which opens new doors for a variety of biological and translational applications. Furthermore, as cell engineering is rapidly developing now, DNA based membrane fusion might become a promising method for cell surface engineering.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (2013CB932803 and 2013CB933800), the National Key R&D Program of China (2016YFA0201200 and 2016YFA0400900), NSFC (21675166, 21390414, 21473236, 31371015 and 21329501), and the Key Research Program of Frontier Sciences, CAS (QYZDJ-SSW-SLH031).
Footnotes
†Electronic supplementary information (ESI) available: Experimental sections, supporting tables and figures. See DOI: 10.1039/c8sc00367j
References
- D’Astolfo D. S., Pagliero R. J., Pras A., Karthaus W. R., Clevers H., Prasad V., Lebbink R. J., Rehmann H., Geijsen N. Cell. 2015;161:674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- Sanchez-Navarro M., Teixido M., Giralt E. Nat. Chem. 2017;9:727–728. doi: 10.1038/nchem.2837. [DOI] [PubMed] [Google Scholar]
- Somia N., Verma I. M. Nat. Rev. Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- Mix K. A., Lomax J. E., Raines R. T. J. Am. Chem. Soc. 2017;139:14396–14398. doi: 10.1021/jacs.7b06597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader B., Baca Q. J., G olan D. E. Nat. Rev. Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- Doudna J. Nature. 2015;528:S6. doi: 10.1038/528S6a. [DOI] [PubMed] [Google Scholar]
- Zuris J. A., Thompson D. B., Shu Y., Guilinger J. P., Bessen J. L., Hu J. H., Maeder M. L., Joung J. K., Chen Z.-Y., Liu D. R. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zuris J. A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q., Georgakoudi I., Liu D. R., Xu Q. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang P., Feng Q., Wang N., Chen Z., Huang Y., Zheng W., Jiang X. NPG Asia Mater. 2017;9:e441. [Google Scholar]
- Stewart M. P., Sharei A., Ding X. Y., Sahay G., Langer R., Jensen K. F. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- Jahn R., Lang T., Sudhof T. C. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Marsden H. R., Tomatsu I., Kros A. Chem. Soc. Rev. 2011;40:1572–1585. doi: 10.1039/c0cs00115e. [DOI] [PubMed] [Google Scholar]
- Xu W., Wang J., Rothman J. E., Pincet F. Angew. Chem., Int. Ed. 2015;54:14388–14392. doi: 10.1002/anie.201506844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Stengel G., Zahn R., Hook F. J. Am. Chem. Soc. 2007;129:9584–9585. doi: 10.1021/ja073200k. [DOI] [PubMed] [Google Scholar]
- Chuard N., Gasparini G., Moreau D., Lörcher S., Palivan C., Meier W., Sakai N., Matile S. Angew. Chem., Int. Ed. 2017;56:2947–2950. doi: 10.1002/anie.201611772. [DOI] [PubMed] [Google Scholar]
- Zong W., Ma S., Zhang X., Wang X., Li Q., Han X. J. Am. Chem. Soc. 2017;139:9955–9960. doi: 10.1021/jacs.7b04009. [DOI] [PubMed] [Google Scholar]
- Kong L., Askes S. H. C., Bonnet S., Kros A., Campbell F. Angew. Chem., Int. Ed. 2016;55:1396–1400. doi: 10.1002/anie.201509673. [DOI] [PubMed] [Google Scholar]
- Yang J., Tu J., Lamers G. E. M., Olsthoorn R. C. L., Kros A. Adv. Healthcare Mater. 2017;6:1700759. doi: 10.1002/adhm.201700759. [DOI] [PubMed] [Google Scholar]
- Torchilin V. P., Rammohan R., Weissig V., Levchenko T. S. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versluis F., Voskuhl J., van Kolck B., Zope H., Bremmer M., Albregtse T., Kros A. J. Am. Chem. Soc. 2013;135:8057–8062. doi: 10.1021/ja4031227. [DOI] [PubMed] [Google Scholar]
- Yang J., Shimada Y., Olsthoorn R. C. L., Snaar-Jagalska B. E., Spaink H. P., Kros A. ACS Nano. 2016;10:7428–7435. doi: 10.1021/acsnano.6b01410. [DOI] [PubMed] [Google Scholar]
- Yang J., Bahreman A., Daudey G., Bussmann J., Olsthoorn R. C. L., Kros A. ACS Cent. Sci. 2016;2:621–630. doi: 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z. J., Yang J., Liu Q., de Vries J. W., Gruszka A., Rodriguez-Pulido A., Crielaard B. J., Kros A., Herrmann A. Chem.–Eur. J. 2017;23:9391–9396. doi: 10.1002/chem.201701379. [DOI] [PubMed] [Google Scholar]
- Zope H. R., Versluis F., Ordas A., Voskuhl J., Spaink H. P., Kros A. Angew. Chem., Int. Ed. 2013;52:14247–14251. doi: 10.1002/anie.201306033. [DOI] [PubMed] [Google Scholar]
- Löffler P. M. G., Ries O., Rabe A., Okholm A. H., Thomsen R. P., Kjems J., Vogel S. Angew. Chem., Int. Ed. 2017;56:13228–13231. doi: 10.1002/anie.201703243. [DOI] [PubMed] [Google Scholar]
- Beales P. A., Vanderlick T. K. Adv. Colloid Interface Sci. 2014;207:290–305. doi: 10.1016/j.cis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Chan Y. H. M., van Lengerich B., Boxer S. G. Proc. Natl. Acad. Sci. U. S. A. 2009;106:979–984. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Gao Y., Xu Y., Chao J., Liu H., Wang L., Li D., Fan C. J. Am. Chem. Soc. 2017;139:17525–17532. doi: 10.1021/jacs.7b09323. [DOI] [PubMed] [Google Scholar]
- Christensen S. M., Bolinger P.-Y., Hatzakis N. S., Mortensen M. W., Stamou D. Nat. Nanotechnol. 2012;7:51–55. doi: 10.1038/nnano.2011.185. [DOI] [PubMed] [Google Scholar]
- Piwonski H. M., Goomanovsky M., Bensimon D., Horovitz A., Haran G. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1437–E1443. doi: 10.1073/pnas.1116670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lengerich B., Rawle R. J., Bendix P. M., Boxer S. G. Biophys. J. 2013;105:409–419. doi: 10.1016/j.bpj.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Rothberg K. G., Anderson R. G. W. J. Cell. Physiol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Li J., Li Q., Huang Q., Shi J. Y., Yan H., Fan C. H. Angew. Chem., Int. Ed. 2014;53:7745–7750. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- Moriyama T., Sasaki K., Karasawa K., Uchida K., Nitta K. J. Cell. Physiol. 2017;232:3565–3573. doi: 10.1002/jcp.25817. [DOI] [PubMed] [Google Scholar]
- Han Y. P., Li X. M., Chen H. B., Hu X. J., Luo Y., Wang T., Wang Z. J., Li Q., Fan C. H., Shi J. Y., Wang L. H., Zhao Y., Wu C. F., Chen N. ACS Appl. Mater. Interfaces. 2017;9:21200–21208. doi: 10.1021/acsami.7b05662. [DOI] [PubMed] [Google Scholar]
- Vancini R., Wang G., Ferreira D., Hernandez R., Brown D. T. J. Virol. 2013;87:4352–4359. doi: 10.1128/JVI.03412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppas N. A., Carr D. A. Chem. Eng. Sci. 2009;64:4553–4565. doi: 10.1016/j.ces.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Shin Y. J. Appl. Polym. Sci. 2007;105:3656–3661. [Google Scholar]
- Shaker M. A., Younes H. M. J. Controlled Release. 2015;217:10–26. doi: 10.1016/j.jconrel.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Akishiba M., Takeuchi T., Kawaguchi Y., Sakamoto K., Yu H.-H., Nakase I., Takatani-Nakase T., Madani F., Graslund A., Futaki S. Nat. Chem. 2017;9:751–761. doi: 10.1038/nchem.2779. [DOI] [PubMed] [Google Scholar]
- Yin P., Choi H. M. T., Calvert C. R., Pierce N. A. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Zhang L., Yang Z., Thu Le T., Teng I. T., Wang S., Bradley K. M., Hoshika S., Wu Q., Cansiz S., Rowold D. J., McLendon C., Kim M.-S., Wu Y., Cui C., Liu Y., Hou W., Stewart K., Wan S., Liu C., Benner S. A., Tan W. Angew. Chem., Int. Ed. 2016;55:12372–12375. doi: 10.1002/anie.201605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bing T., Shen L., Song R., Wang L., Liu X., Liu M., Li J., Tan W., Shangguan D. Angew. Chem., Int. Ed. 2016;55:3914–3918. doi: 10.1002/anie.201510786. [DOI] [PubMed] [Google Scholar]
- Seo S. E., Li T., Senesi A. J., Mirkin C. A., Lee B. J. Am. Chem. Soc. 2017;139:16528–16535. doi: 10.1021/jacs.7b06734. [DOI] [PubMed] [Google Scholar]
- Wan S., Zhang L., Wang S., Liu Y., Wu C., Cui C., Sun H., Shi M., Jiang Y., Li L., Qiu L., Tan W. J. Am. Chem. Soc. 2017;139:5289–5292. doi: 10.1021/jacs.7b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Fu T., Wang R., Liu H., Zou J., Zhao Z., Ye M., Zhang X., Tan W. Chem. Sci. 2017;8:7082–7086. doi: 10.1039/c7sc02819a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Shen J., Chen S., Wang D., Li F., Mathur S., Song S., Fan C. Chem. Sci. 2014;5:4460–4466. [Google Scholar]
- Wang S., Cai X., Wang L., Li J., Li Q., Zuo X., Shi J., Huang Q., Fan C. Chem. Sci. 2016;7:2722–2727. doi: 10.1039/c5sc04102c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Z., Weber T. J., Hu D., Lin C.-T., Li J., Lin Y. Anal. Chem. 2013;85:6775–6782. doi: 10.1021/ac400858g. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Qiao Y., Hao F., Zhang L., Wu S., Li Y., Li J., Song X.-M. Chem.–Eur. J. 2010;16:8133–8139. doi: 10.1002/chem.201000684. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang L., Li Z., Lin Y., Li J. Nat. Protoc. 2014;9:1944–1955. doi: 10.1038/nprot.2014.126. [DOI] [PubMed] [Google Scholar]
- Lutolf M. P., Weber F. E., Schmoekel H. G., Schense J. C., Kohler T., Muller R., Hubbell J. A. Nat. Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- Richardson T. P., Peters M. C., Ennett A. B., Mooney D. J. Nat. Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- Battig M. R., Soontornworajit B., Wang Y. J. Am. Chem. Soc. 2012;134:12410–12413. doi: 10.1021/ja305238a. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y., Winfree E. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- Dirks R. M., Pierce N. A. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z. L., Lin M. H., Wang P., Pei H., Yan J., Sho J. Y., Huang Q., He D. N., Fan C. H., Zuo X. L. Anal. Chem. 2014;86:2124–2130. doi: 10.1021/ac4037262. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wang H., Jiang J.-H., Shen G.-L., Yu R.-Q., Li J. Anal. Chem. 2009;81:1982–1987. doi: 10.1021/ac802512d. [DOI] [PubMed] [Google Scholar]
- Jiang X. J., Wang X. D. J. Biol. Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Jiang X. J., Wang X. D. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- Dutta K., Hu D., Zhao B., Ribbe A. E., Zhuang J. M., Thayumanavan S. J. Am. Chem. Soc. 2017;139:5676–5679. doi: 10.1021/jacs.7b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam N. W. S., Dai H. J. J. Am. Chem. Soc. 2005;127:6021–6026. doi: 10.1021/ja050062v. [DOI] [PubMed] [Google Scholar]
- Lee H., Dam D. H. M., Ha J. W., Yue J., Odom T. W. ACS Nano. 2015;9:9859–9867. doi: 10.1021/acsnano.5b05138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Lv J., Gao X., Wang X., Wang H., Chen H., He X., Li L., Cheng Y. Nano Lett. 2017;17:1678–1684. doi: 10.1021/acs.nanolett.6b04955. [DOI] [PubMed] [Google Scholar]
- Kim H., Shin K., Park O. K., Choi D., Kim H. D., Baik S., Lee S. H., Kwon S.-H., Yarema K. J., Hong J., Hyeon T., Hwang N. S. J. Am. Chem. Soc. 2018;140:1199–1202. doi: 10.1021/jacs.7b08440. [DOI] [PubMed] [Google Scholar]
- Capicciotti C. J., Zong C., Sheikh M. O., Sun T., Wells L., Boons G.-J. J. Am. Chem. Soc. 2017;139:13342–13348. doi: 10.1021/jacs.7b05358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kwong B., Irvine D. J. Angew. Chem., Int. Ed. 2011;50:7052–7055. doi: 10.1002/anie.201101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.