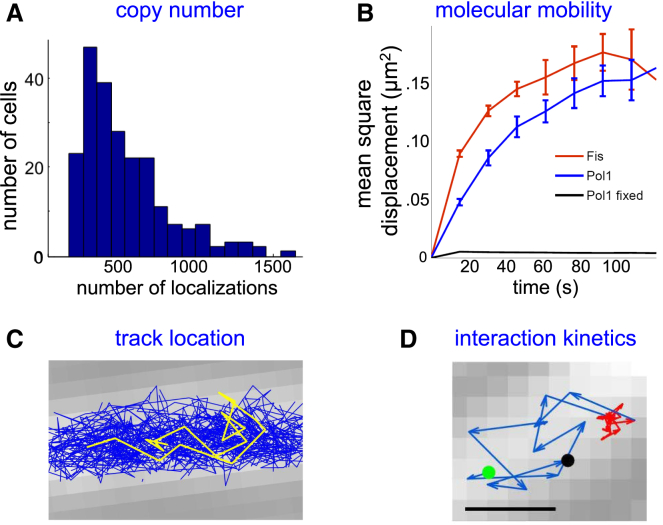
Figure 4.
Main single-molecule fluorescence observables inside living bacteria using fluorescent proteins. The observables apply for both autofluorescent and photoactivatable proteins, although the latter will provide higher statistics for counting and tracking. (A) The number of localizations per cell is shown, corresponding loosely to the protein copy number per cell. (B) Molecular mobility can be examined using plots of mean-square displacement (MSD) (pictured) or the cumulative distribution function (which plots the cumulative probability of finding a molecule within a certain distance after a certain time); this information can also be converted into apparent diffusion coefficients per track. In the example, a DNA polymerase (Pol1) in fixed cells shows no significant motion, whereas in live cells, it shows significant displacements until the confinement effects cause saturation of the MSDs; a smaller DNA-binding protein, Fis, shows faster motion. (C) Track location can be examined relative to the cell boundaries, relative to all other tracks (as pictured), relative to tracks with the same or different mobility, and relative to cell landmarks monitored in a different detection channel. (D) Time-series analysis of individual tracks can provide information about interaction (binding) kinetics identified by changes in the molecular mobility. In the example, a DNA polymerase molecule identifies its target, performs DNA synthesis, and resume its target search. Each step is 15 ms. Scale bars, 500 nm. The example figures are taken from (50). To see this figure in color, go online.