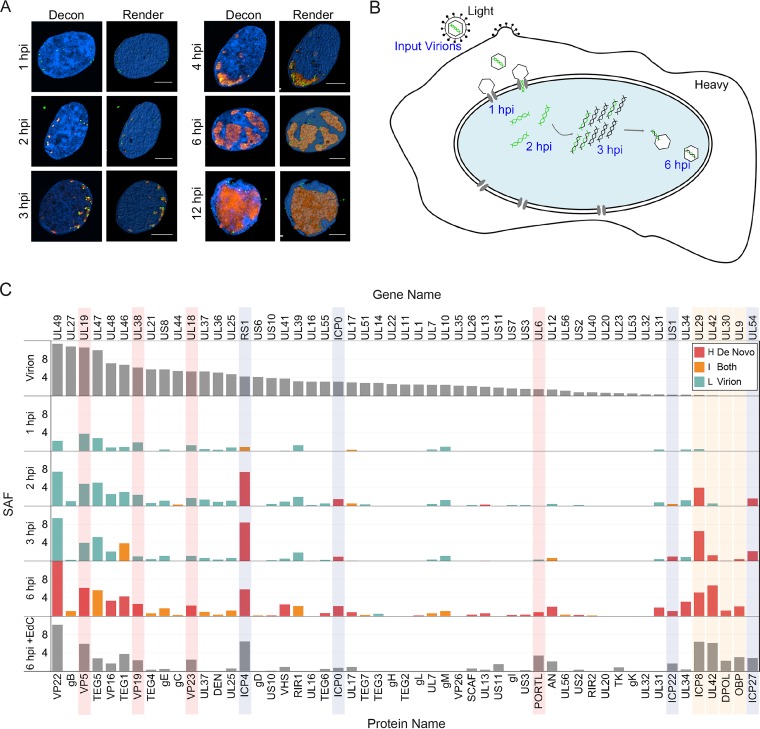
FIG 1 .
The fates of input viral genomes can be tracked within infected-cell nuclei. (A) Visualization of input viral DNA. Cells were fixed at the indicated times after infection with KOS-EdC (1 to 12 hpi) at an MOI of 10 PFU/cell. Viral genomes (green) and ICP4 (red) were imaged relative to host nuclei (blue). Optical sections were deconvolved (Decon) to generate high-resolution images and rendered (Render) to illustrate the positions of input viral DNA. Bars, 5 µm. (B) Model depicting the fate of input viral genomes during distinct stages of infection. Input viral DNA is shown in green, and replicated viral DNA is shown in black. For MS experiments, input virions and the infected cell contain proteins labeled with light or heavy amino acids, respectively. (C) Abundance of viral-genome-associated viral proteins throughout infection. Viral-genome-associated viral proteins were detected by MS after a 1-, 2-, 3-, or 6-h infection with KOS-EdC (MOI of 10 PFU/cell). Spectral abundance factors (SAFs) were plotted relative to individual proteins associated with mature virions (Virion) and viral replication compartments that were labeled with EdC from 4 to 6 hpi (6 hpi + EdC) (15). Proteins were distinguished as either heavy (H), light (L), or intermediate (I) by SILAC analysis. In cases where SILAC analysis was not carried out, bar graphs are shown in gray. IE viral proteins are shown on a light gray background, viral replication proteins on tan background, and capsid proteins on pink background. Data represent results from one of two biological replicates. See also Tables S1 and S2 and Fig. S1 and S2 in the supplemental material.