Figure 13. Structural deviations and residue-residue contact score:
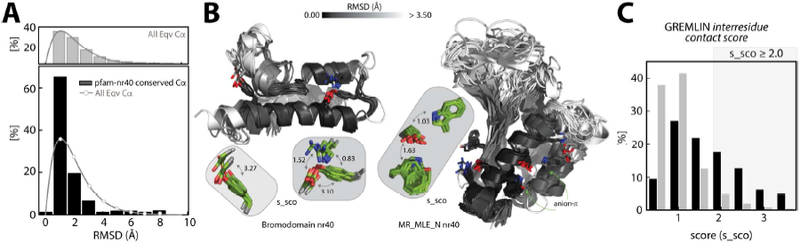
(A) Root-mean-square deviation (RMSD) distribution (top, gray bars) of all equivalent Cα atoms in the structural alignment of pfam_nr40 structures. The RMSD distribution of Cα atoms (bottom, black bars) of conserved interacting positions (anion-quadrupole, HB-anion–quadrupole, and salt bridge) in the pfam_nr40 structures. The background distribution (gray line) of all equivalent Cα atoms is overlaid. (B) Structural deviation by RMSD (white, ≥ 3.5 Å and black, 0.0 A) in the pfam_nr40 structures of the bromodomain (left) and MR_MLE_N (right) domain. Anion–quadrupole-interacting positions in these structures are also shown in the inset in green with their GREMLIN s_sco scores indicated with curved, double-headed arrows. (C) The distribution of the s_sco scores (black bars) for positions conserved in at least two of the pfam_nr40 structures. Gray bars represent the background distribution of the GREMLIN s_sco scores in the dataset.
