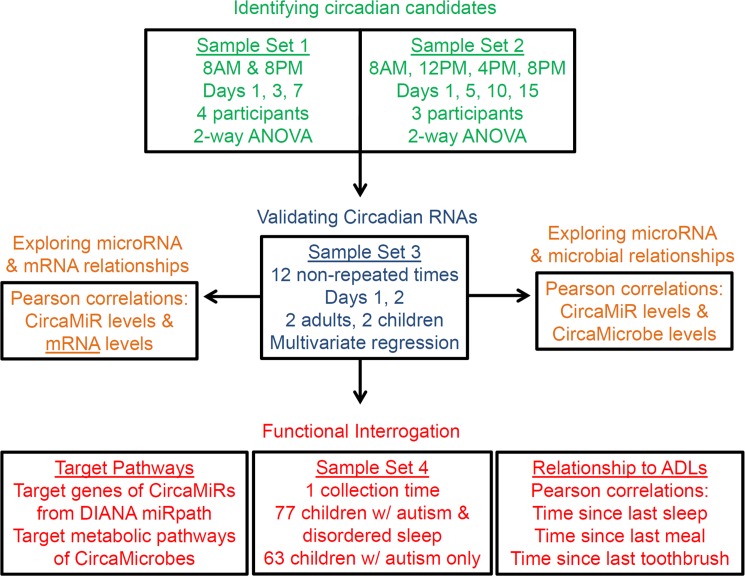
Fig 1. Flow chart outlining the analytic approach for the current study.
Briefly, sample sets 1 and 2 were used to identify circadian RNA candidates (green), which were then validated in sample set 3 (blue). Relationships between CircaMiR levels and CircaMicrobes, or mRNA targets were explored (orange). The functional implications of CircaMiRs and CircaMicrobes were interrogated through annotation analyses and characterization in a cohort of children with disordered sleep (sample set 4; red). The relationship of oscillating RNA with patterns of daily activity (sleep, eating, and tooth brushing) were also investigated.