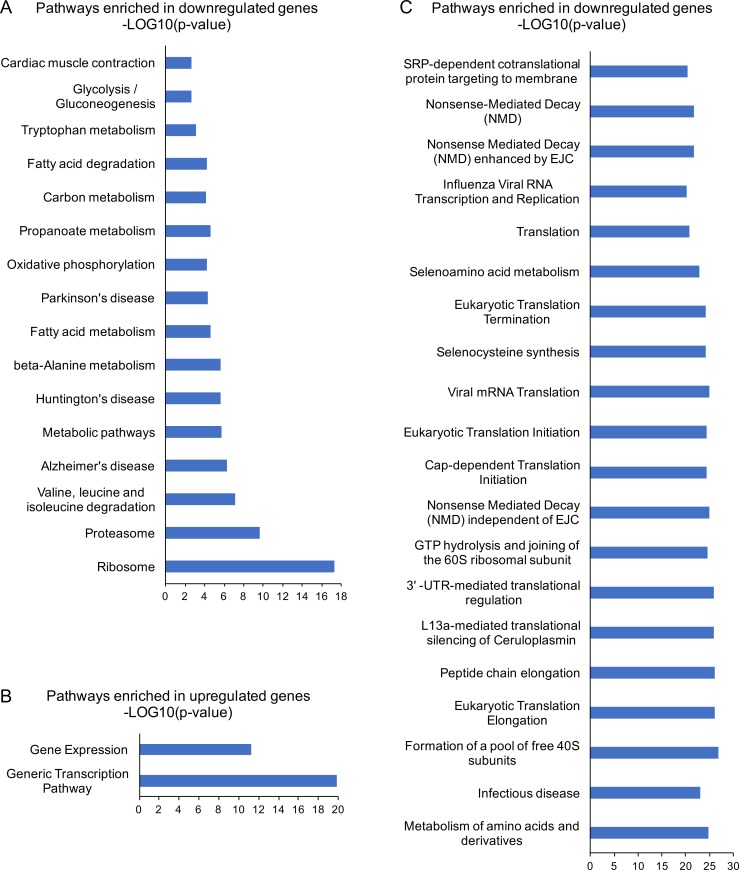
Fig 2. Gene set enrichment analysis of Met/Cys-deprived HeLa cells.
Differentially expressed genes between three time points of starvation (15-30-72 h) and controls were selected based on a P value <0.05 and a fold change of at least 2, leading to a total of 996 upregulated, and 1037 downregulated genes. The enrichment analysis was performed separately for up and down regulated genes, using the EnrichR tool and the KEGG (A) and REACTOME (B, C) databases. Ranking is based on the combined score provided by EnrichR, and categories are displayed up to 20 items with an Adjusted P value <0.05. No significant categories were found with upregulated genes against the KEGG database. All data are shown in S1 File. The enrichment analysis using all differentially expressed genes together did not reveal any additional enriched process.