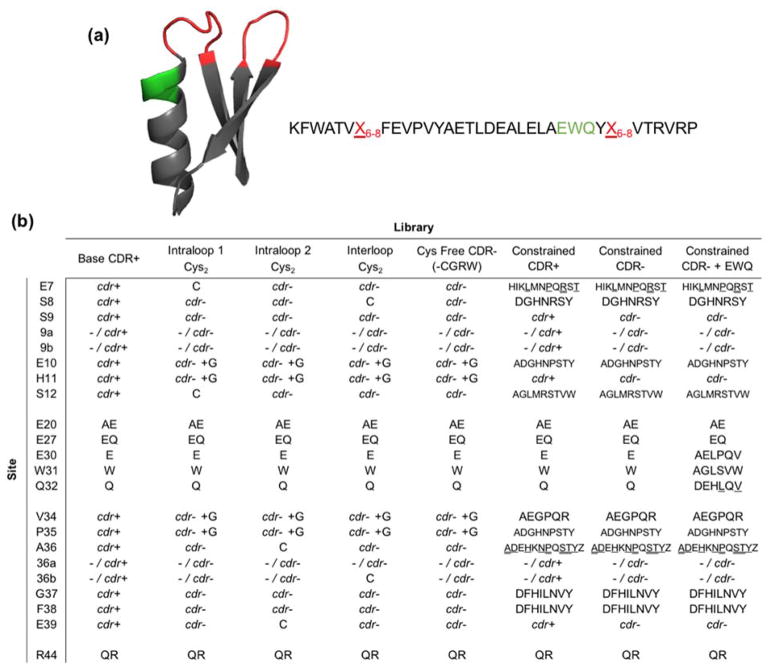
Figure 1. Gp2 protein scaffold structure and second generation library designs.
(a) Cartoon ribbon structure and amino acid sequence of the Gp2 scaffold (PDB ID: 2WNM). The paratope region (red) is allowed to remain wild-type length or extend by one or two amino acids. A region near paratope loop two that previously displayed higher than expected mutation rates is shown in green. (b) Combinatorial library design: codon or amino acid diversity for each site in each library. cdr+ represents base antibody CDR diversity and cdr− represents the closest match to that diversity while removing cysteine (and thus arginine, glycine, and tryptophan due to degenerate codon limitations, except when glycine is added on a separate codon (cdr− +G)). Single amino acid abbreviations are used for diversity at constrained sites, at approximately equal frequency except where denoted by underline (higher) or double underline (highest). Full details in Supplemental Tables 1 and 2.