Abstract
Molecular cues from environmental bacteria influence important developmental decisions in diverse marine eukaryotes. Yet, relatively little is understood about the mechanisms underlying these interactions, in part because marine ecosystems are dynamic and complex. With the help of simple model systems, including the choanoflagellate Salpingoeca rosetta, we have begun to uncover the bacterial cues that shape eukaryotic development in the ocean. Here, we review how diverse bacterial cues — from lipids to macromolecules — regulate development in marine eukaryotes. It is becoming clear that there are networks of chemical information circulating in the ocean, with both eukaryotes and bacteria acting as nodes; one eukaryote can precisely respond to cues from several diverse environmental bacteria, and a single environmental bacterium can regulate the development of different eukaryotes.
Introduction
Eukaryotes evolved over two billion years ago in a world dominated by prokaryotes and have lived in close association with bacteria ever since. It has become increasingly clear that bacteria not only act as competitors and pathogens, but also promote proper health and development in eukaryotes [1,2]. Growing attention has focused on how the microbiome shapes many aspects of eukaryotic development, from root nodule development in legumes [3], to light organ morphogenesis in the Hawaiian bobtail squid [4], and even immune system development in vertebrates [5]. Yet, bacteria in the microbiome are not the only bacteria influencing eukaryotic development. Although often overlooked, free-living environmental bacteria also provide cues that regulate essential developmental processes in diverse eukaryotes.
Many examples of interactions between environmental bacteria and eukaryotes stem from marine ecosystems, where bacterial cues elicit developmental transitions in organisms as diverse as algae and animals. One of the best explored examples concerns the mutualistic interaction between the Hawaiian bobtail squid (Euprymna scolopes) and the bacterium Vibrio fischeri, in which colonization of the squid by environmental V. fischeri is required to induce the morphogenesis of the host’s light-organ (Table 1) [4]. The squid-Vibrio association is unusually amenable to laboratory manipulation, and as such, many of the relevant bacterial cues have been identified and their consequences for squid development explored in detail [6–9]. In contrast, few other marine bacterial-eukaryotic interactions are understood in molecular detail, in part because marine environments are host to dynamic and diverse bacterial communities. While it is challenging to decipher specific interactions in such complex ecosystems, the lessons learned by exploring diverse marine host-microbe interactions are likely to extend to interactions between eukaryotes and other bacterial communities, such as those in the gut and soil.
Table 1.
Environmental bacteria regulate development in marine eukaryotes
| Eukaryote | Bacteria (Phylum) | Developmental outcome | Molecular cue | Reference |
|---|---|---|---|---|
| Chlorophyta (green algae) | ||||
| Ulva mutabilis | Cytophaga (B) | Thallus differentiation | Unknown | [32•] |
| Roseobacter (B) | Cell division | Unknown | [32•] | |
| Algoriphagus (B) | Cell division and thallus differentiation | Unknown | [42,43] | |
| Polaribacter (B) | Cell division and thallus differentiation | Unknown | [42,43] | |
| Monostroma oxyspermum | Zobellia uliginosa (B) | Morphogenesis | Thallusin | [30••] |
| Ulva pertussa | Zobellia uliginosa (B) | Morphogenesis | Unknown | [29] |
| Ulva conglobata | Zobellia uliginosa (B) | Morphogenesis | Unknown | [29] |
| Ulva fasciata | Marinomonas (F) | Morphogenesis/growth of zoospores | Unknown | [44] |
| Bacillus (F) | Morphogenesis/growth of zoospores | Unknown | [44] | |
| Enteromorpha | Vibrio anguillarum (G) | Zoospore settlement | AHLs | [45] |
| Choanoflagellate | ||||
| Salpingoeca rosetta | Algoriphagus machipongonensis (B) | Rosette development | Lipid cofactors (Sulfonolipids, LPEs, capnine) | [15,16••,17] |
| Zobellia uliginosa (B) | Rosette development | Uncharacterized lipids | [15]; unpublished data |
|
| Demequina (A) | Rosette development | Uncharacterized lipids | [15]; unpublished data |
|
| Vibrio fischeri (G) | Sexual reproduction | Chondroitin lyase | [22••] | |
| Vibrio orientalis (G) | Sexual reproduction | Unknown | [22••] | |
| Vibrio tubiashii (G) | Sexual reproduction | Unknown | [22••] | |
| Flavobacterium heparinum (B) | Sexual reproduction | Chondroitin lyase | [22••] | |
| Porifera (sponges) | ||||
| Rhopaloeides odorabile | Bacterial biofilm | Larval settlement | Unknown | [46] |
| Cnidaria | ||||
| Acropora millepora (coral) | Pseudoalteromonas (G) | Larval metamorphosis | Tetrabromopyrrole | [34••] |
| Acropora willisae (coral) | Pseudoalteromonas (G) | Larval metamorphosis | Tetrabromopyrrole | [44] |
| Porites astreoides (coral) | Pseudoalteromonas (G) | Larval metamorphosis | Tetrrabromopyrrole | [44] |
| Cassiopea andromeda (jellyfish) | Vibrio alginolyticus (G) | Larval metamorphosis | Unknown | [47] |
| Aurelia aurita (jellyfish) | Micrococcacae (A) | Larval settlement | Glycolipids | [48] |
| Mollusca | ||||
| Euprymna scolopes (Hawaiian bobtail squid) | Vibrio fischeri (G) | Light organ morphogenesis | Lipopolysaccharide (LPS); Peptidoglycan (PGN); Tracheal Cytotoxin (TCT) | [7–9,49] |
| Crassostrea gigas (oyster) | Altermonas colwelliana (G) | Larval settlement/ metamorphosis | Unknown | [50] |
| Vibrio cholerae (G) | Larval settlement/ metamorphosis | Unknown | [50] | |
| Annelida | ||||
| Hydroides elegans (marine tubeworm) | Pseudoalteromonas luteoviolacea (G) | Larval settlement | Tailocin MACs | [35••] |
| Cellulophaga lytica (B) | Larval settlement | Unknown | [51•] | |
| Bacillus aquimaris (F) | Larval settlement | Unknown | [51•] | |
| Staphylococcus warneri (F) | Larval settlement | Unknown | [51•] | |
| Echinodermata | ||||
| Heliocidaris erythrogramma (sea urchin) | Pseudoalteromonas luteoviolacea (G) | Larval settlement | Unknown | [52] |
| Vibrio (G) | Larval settlement | Unknown | [52] | |
| Shewanella (G) | Larval settlement | Unknown | [52] | |
| Chordata | ||||
| Ciona intestinalis (sea squirt) | Pseudomonas (G) | Larval attachment | Exopolysaccharide | [53] |
Bacterial phylogeny key: (B) Bacteroidetes; (G) Gammaproteobacteria; (F) Firmicutes; (A) Actinobacteria.
Simple model systems are beginning to reveal how environmental bacteria shape eukaryotic development in the ocean. Important features of these models that facilitate the identification of molecules underlying bacterial-eukaryotic interactions include: (1) the ability to grow and manipulate both the bacteria and the eukaryote in the lab, and (2) a clear and quantifiable response of the eukaryote to a single bacterium. Here we review mechanisms by which environmental bacteria regulate the development of choanoflagellates and other marine eukaryotes to illustrate how, and explore why, important eukaryotic developmental decisions rely on cues from specific environmental bacteria.
A choanoflagellate model for bacterial-eukaryotic interactions
One of the closest living relatives of animals, the choano-flagellate Salpingoeca rosetta, has emerged as an attractive model for investigating how environmental bacteria shape eukaryotic cell biology and life history. Choano-flagellates are unicellular and colony-forming microeukaryotes that live in diverse aquatic environments [10]. Every choanoflagellate cell bears an apical ‘collar complex’ — a single flagellum surrounded by a feeding collarcomposed of actin-filled microvilli — that it uses to capture and phagocytose bacterial prey. Importantly, the collar complex and its role in mediating interactions with bacteria are conserved among choanoflagellates and animals [10–12]. However, choanoflagellates do not just eat bacteria, but they also undergo key life history transitions in response to molecular cues secreted by environmental bacteria.
A network of bacterial lipids flips a developmental switch in S. rosetta
In many choanoflagellates, including the emerging model choanoflagellate S. rosetta, a solitary cell can develop into a multicellular ‘rosette’ colony through serial rounds of oriented cell division, with the sister cells remaining stably adherent [13,14] (Figure 1a). Although S. rosetta was isolated from the ocean as a rosette, early laboratory cultures proliferated primarily in the unicellular form, producing rosettes infrequently and unpredictably. A set of unexpected observations revealed that Algoriphagus machipongonensis, an environmental bacterium that had been co-isolated with the choanoflagellate and persisted in laboratory cultures at very low densities, could induce consistent and uniform rosette development in S. rosetta when grown at higher densities [15].
Figure 1.
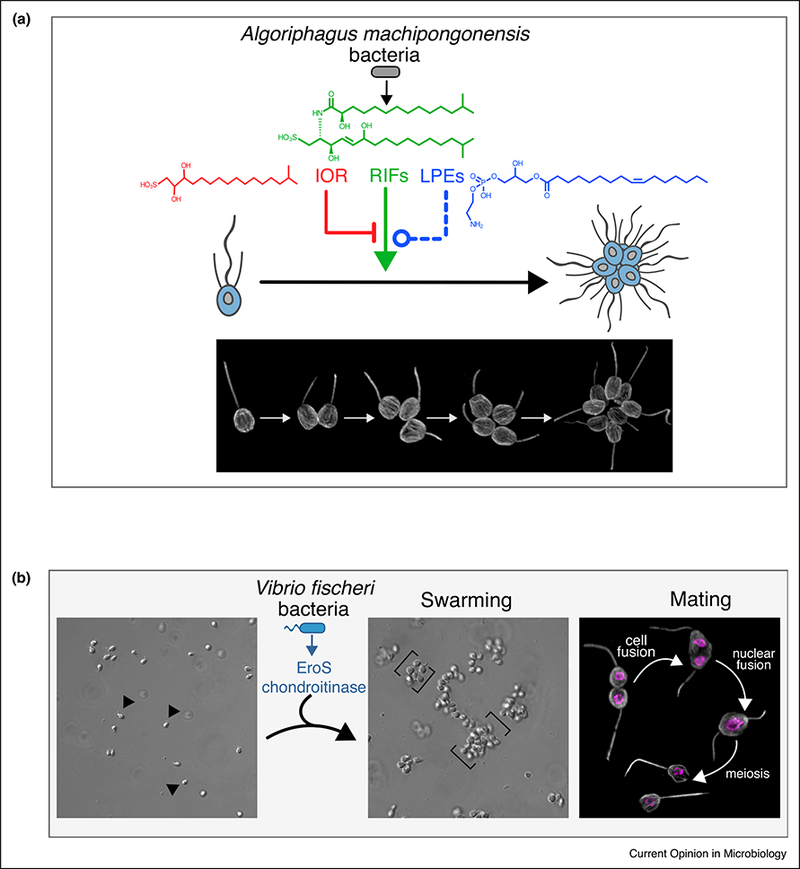
Bacteria regulate rosette development and sexual reproduction in the choanoflagellate, S. rosetta. (a) Algoriphagus machipongonensis bacteria regulate the development of S. rosetta from a solitary cell into a multicellular ‘rosette’ colony through serial rounds of cell division. Algoriphagus produces three classes of lipids — sulfonolipids (RIFs), lysophosphatidylethanolamines (LPEs), and a capnine (IOR-1) — that interact to alternately induce, enhance, or inhibit rosette development. While the sulfonolipid RIFs are sufficient to initiate rosette development in S. rosetta, they require the synergistic enhancing activity of the LPEs for robust rosette development. Algoriphagus also produces the inhibitory IOR-1 that inhibits the RIFs, but cannot overcome the synergistic inducing activity of the RIFs + LPEs. Immunofluorescence images illustrate stages of S. rosetta rosette development; tubulin staining (gray) highlights the cell body and apical flagellum. (b) Vibrio fischeri bacteria induce sexual reproduction in S. rosetta. EroS, a chondroitin lyase secreted by V. fischeri, triggers solitary S. rosetta cells (arrows) to form large swarms (brackets) through cell aggregation. During swarming, S. rosetta cells pair off and mate, a process that involves the cell and nuclear fusion of two haploid cells into one diploid cell, followed by meiosis to generate haploid progeny. Immunofluorescence images depict mating stages in S. rosetta; tubulin staining (gray) highlights the cell body and apical flagellum, and Hoechst staining (magenta) highlights the nucleus.
Because S. rosetta and Algoriphagus could be cultured independently or together, and because rosette development was quantifiable (i.e. % of cells in rosettes), a straightforward rosette development bioassay could be used to investigate the molecular basis of Algoriphagus rosette-inducing activity. Activity-guided fractionation led to the isolation of RIF-1 (Rosette-Inducing Factor-1), a novel sulfonolipid signaling molecule that induced rosette development in S. rosetta [15]. However, only a small fraction of S. rosetta cells formed rosettes in response to RIF-1, far fewer than that induced by live Algoriphagus, leading to the hypothesis that additional Algoriphagus molecules influence S. rosetta rosette development [16••].
Further work revealed that Algoriphagus produces additional lipid activators, synergistic enhancers, and inhibitors that regulate rosette development [16••,17] (Figure 1a). While the RIFs (RIF-1 and a second sulfo-nolipid, RIF-2) were sufficient to induce low levels of rosette development, an additional class of lipid synergists, the lysophosphatidylethanolamines (LPEs), were required for robust rosette induction. Together, the RIFs and LPEs recapitulated the full rosette inducing activity of live Algoriphagus.
The importance of the LPEs had initially been obscured by the fact that they did not exhibit any bioactivity on their own; only by testing bacterial lipid fractions in combination with the RIFs did it become clear that these synergistic lipids helped to fully potentiate the induction of rosette development. Testing bacterial fractions in combination also revealed that Algoriphagus produces a molecule that competes with and inhibits RIF-induced rosette development. The molecule, a capnine called IOR-1 (Inhibitor of Rosettes-1), antagonizes the RIFs, but its inhibitory activity can be bypassed in the presence of LPEs, providing a possible explanation for why IOR-1 does not normally prevent Algoriphagus rosette induction.
It is interesting to contemplate what evolutionary processes might have led S. rosetta to rely upon a network of bacterial cues before committing to rosette development. We hypothesize that certain bacteria serve as proxies for environmental conditions; over time, the stochastic process of natural selection may have favored individuals that required multiple bacterial cues before initiating rosette development, thus avoiding making the switch to rosette development under unfavorable environmental conditions. This integrated response may be especially important in aquatic environments, where bacterial composition and nutrient availability are constantly changing.
A bacterial chondroitinase triggers mating in S. rosetta
In addition to rosette development, S. rosetta can transition from asexual proliferation to sexual reproduction, wherein solitary haploid cells fuse to produce a diploid cell that will then undergo meiosis [18]. Despite harboring a complete meiotic genetic toolkit [19,20], the S. rosetta sexual cycle was rarely observed in laboratory cultures. Only under starvation conditions would a small fraction of the S. rosetta population mate [21]. A serendipitous observation revealed that specific environmental bacteria, missing from most laboratory cultures, were capable of triggering a population-wide switch to sexual reproduction [22••].
This discovery stemmed from the observation that Vibrio fischeri, an abundant marine bacterium, induced the formation of large motile aggregates or ‘swarms’ composed of many solitary S. rosetta cells. Swarming had not been previously described in S. rosetta, and further examination revealed that during swarming, haploid S. rosetta cells frequently paired off and underwent cell and nuclear fusion. Genetic experiments confirmed that the diploid products of cell and nuclear fusion later generated meiotic progeny, demonstrating that V. fischeri bacteria induce the full sexual cycle in S. rosetta (Figure 1b). We subsequently found that Vibrio orientalis and Vibrio tubiashii elicit swarming and mating in S. rosetta, while other species tested did not, suggesting that S. rosetta mating in nature might be regulated by some, but not all species of Vibrio bacteria [22••].
Because swarming was always observed prior to mating, S. rosetta swarming provided a robust bioassay for identifying the molecular basis of the V. fischeri ‘aphrodisiac’ activity. Activity-guided fractionation led to the isolation of a protein, named EroS (Extracellular Regulator of Sex) that fully recapitulated the activity of Vibrio bacteria. Biochemical assays revealed that EroS belongs to a class of bacterial polysaccharide-degrading enzymes called chondroitinases, and that the chondroitin-degrading activity of EroS is sufficient to induce mating in S. rosetta. Finally, the S. rosetta target of EroS was identified as the sulfated polysaccharide chondroitin sulfate, a component of the extracellular matrix previously thought to be restricted to the animal lineage. As the first example of an environmental bacterium regulating eukaryotic sexual reproduction, the interaction between V. fischeri and S. rosetta raises the possibility that mating in other aquatic eukaryotes may be influenced by environmental bacteria as well.
Bacteria as master regulators of S. rosetta life history in the marine environment
Bacteria are required for rosette development and mating under laboratory conditions — but can bacteria plausibly regulate S. rosetta development in nature? Despite their underlying molecular differences, the cues that induce rosette development and mating are bioactive at environmentally relevant concentrations. The purified Algoripha-gus RIFs and LPEs display activity at high nanomolar to low micromolar concentrations in the laboratory; yet, the hydrophobicity of these molecules makes it unlikely that S. rosetta encounters them as isolated lipids in the environment. As constituents of the Algoriphagus outer membrane, it is likely that RIFs and LPEs are packaged into outer membrane vesicles (OMVs), spherical packages of periplasmic content constitutively produced by Gram negative bacteria [23,24], and thereby released into the environment. Indeed, Algoriphagus OMVs elicit robust rosette development [16••], and retain their bioactivity under a wide range of conditions. Moreover, diverse bacteria belonging to several marine Bacteroidetes and Actinobacteria genera induce rosette development in S. rosetta ([15]; unpublished data]), raising the likelihood that S. rosetta might encounter rosette-inducing bacteria in multiple environments
In contrast with the lipid regulators of rosette development, the mating-inducing chondroitin lyase, EroS, is a soluble protein constitutively secreted by V. fischeri bacteria. Not only does EroS trigger mating at picomolar concentrations, but S. rosetta swarms in response to as few as 400 V. fischeri cells/mL — a density similar to that of V. fischeri in oligotrophic oceans [25]. In addition to V. fischeri, other species of Vibrio bacteria also induce mating in S. rosetta (including V. orientalis and V. tubiashii), as do commercial chondroitin lyases isolated from Flavobacterium heparinum and Proteus vulgaris [22••], suggesting that encounters between S. rosetta and molecules produced by mating-inducing bacteria might be common occurrences in the ocean.
Thus, it is reasonable to infer that S. rosetta comes across both rosette development and mating inducing bacteria in nature. In addition, because S. rosetta can respond to cues from diverse bacteria, it seems plausible that other life history transitions in choanoflagellates, such as settlement (the attachment of a planktonic cell to a substrate; [10,14]), are regulated by environmental bacteria as well.
The widespread influences of environmental bacteria
Choanoflagellates are not the only eukaryotes taking life advice from environmental bacteria. Environmental bacteria also regulate developmental transitions in diverse marine algae and animals, and simple model systems are beginning to uncover the bacterial cues that influence eukaryotic morphogenesis (Figure 2).
Figure 2.
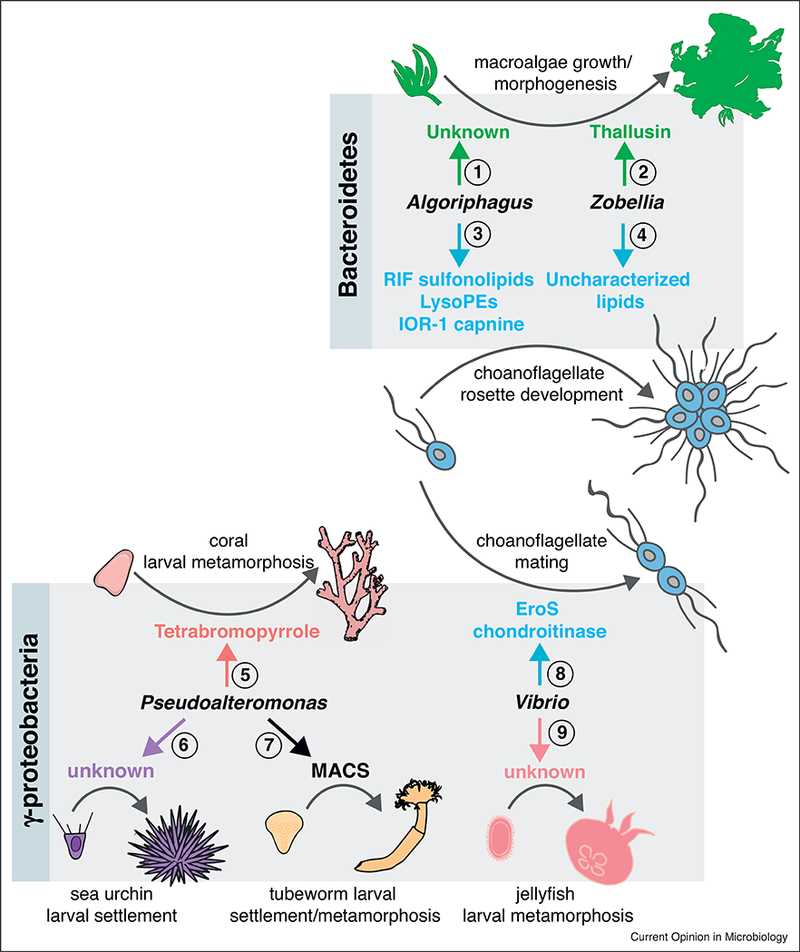
Distinct molecular cues from environmental Bacteroidetes and Gammaproteobacteria regulate developmental transitions in diverse marine eukaryotes. The Bacteroidetes bacteria Algoriphagus and Zobellia uliginosa regulate morphogenesis in organisms as diverse as algae and choanoflagellates. (1) Uncharacterized factors produced by Algoriphagus induce morphogenesis in the macroalgae Ulva mutabilis. (2) Algoriphagus machipongonensis lipids [sulfonolipids, lysophosphatidylethanolamines, and a capnine] regulate rosette development in the choanoflagellate Salpingoeca rosetta. (3) Thallusin, an amino acid derivative produced by Zobellia uliginosa, induces morphogenesis in the macroalgae Monostroma oxyspermum. (4) Uncharacterized molecules from Zobellia uliginosa induce rosette development in the choanoflagellate Salpinogeca rosetta. Gammaproteobacteria can likewise elicit developmental responses in diverse animals and choanoflagellates. (5) Tetrabromopyrrole produced by Pseudoalteromonas spp. induces larval metamorphosis in corals Acropora millepora and Acropora willisae, and larval settlement (attachment and metamorphosis) in the coral Porites astreoides. (6) Uncharacterized cues from Pseudoalteromonas bacteria induce larval settlement in the sea urchin Heliocidaris erythrogramma. (7) Pseudoalteromonas luteoviolacea produces arrays of contractile phage tail-like structures (MACs) that trigger metamorphosis of the tubeworm Hydroides elegans. (8) A chondroitinase (EroS) secreted by Vibrio fischeri induces mating in the choanoflagellate S. rosetta. (9) Unknown cues secreted by Vibrio alginolyticus induce larval metamorphosis in the jellyfish Cassiopea andromeda.
Algal morphogenesis
It has long been known that microbial communities associated with the surfaces of marine macroalgae are essential for their growth and morphogenesis [26]. Yet, the bacterial species responsible for stimulating algal development remained elusive for many years, due to the complex and seasonally-shifting composition of algal-associated bacterial communities [27]. A key advance in studying bacterial-algal interactions was the development of axenic culturing techniques (including for the seaweed Monostroma oxyspermum), which provided a platform for testing individual bacterial species for morphogenesis-inducing activity [28]. Although diverse bacteria that influence algal growth and morphogenesis have now been identified (Table 1), only one morphogenetic factor, Thallusin, has been isolated by activity-guided fractionation and characterized to date [29,30••,31]. Thallusin is an amino acid derivative produced by the Monostroma- associated bacterium, Zobellia uliginosa, that is sufficient to induce thallus development in M. oxyspermum and partially promote thallus development in Ulva species (Figure 2). With a clear bioassay available, why have not more algal morphogenetic factors been isolated? One hypothesis is that the bacterial cues regulating algal growth and morphogenesis are produced at very low levels. This was certainly true for Thallusin, although it was ultimately possible to isolate the molecule because of its potency and stability [30••]. Alternatively, induction may require multiple bacterial molecules. On their own, bacteria belonging to Cytophaga and Roseobacter genera induce incomplete Ulva mutabilis development, promoting either cell division or thallus differentiation, respectively [32•]. However, the combined activities of these bacteria fully restore normal morphogenesis, raising the possibility that the synergistic interactions observed at the organismal level are required at the molecular level as well.
Larval settlement
Many benthic marine invertebrates have complex life histories that include stages of larval settlement and metamorphosis, key developmental steps that are crucial for adult success. Bacterial biofilms provide cues that trigger larval settlement and metamorphosis in diverse marine invertebrates, including sponges, cnidarians, molluscs, annelids, echinoderms, and urochordates (Table 1). While the majority of these interactions remain poorly understood, the increased tractability of invertebrate systems (e.g. the tubeworm Hydroides elegans [33] and the coral Acropora millepora [34••]) has facilitated identification of the bacterial cues that regulate larval settlement and metamorphosis. Biofilm-forming bacteria from the genus Pseudoalteromonas influence development in several animals, including corals and tubeworms. Interestingly, the cues produced by environmental Pseudoalteromonas that trigger metamorphosis in A. millepora and H. elegans are distinct; while A. millepora metamorphosis is induced by the small molecule tetrabromopyrrole [34••], H. elegans metamorphosis is regulated by arrays of contractile phage-tail like structures called MACs (Metamorphosis Associated Contractile Structures) [35••] (Figure 2). Although the structures of tetrabromopyrrole and MACs suggest that the mechanisms by which these molecules trigger metamorphosis are likely very different, both of these bacterial molecules may provide chemical evidence of a suitable surface for colonization. Because surfaces in the ocean are often limiting, cues from bacteria might indicate to animals that they have found an appropriate environment for settling down.
Bacterial cues are proxies for environmental conditions
As more bacterial cues are isolated, it is becoming clear that interactions between eukaryotes and their environmental bacteria exhibit remarkable molecular specificity. Even slight modifications to the structures of bacterial cues can completely eliminate inducing activity, as is the case with the choanoflagellate rosette-inducing molecules and the algal morphogenetic factor Thallusin [17,31,36]. Nonetheless, multiple environmental bacteria can elicit the same eukaryotic developmental responses, and in each case the molecular cues seem to be distinct (this has been demonstrated for S. rosetta rosette development, Monostroma morphogenesis, and H. elegans larval settlement; Table 1). Because marine microbial communities are highly dynamic, it may be beneficial for eukaryotes to interpret developmental cues from diverse bacteria. The molecular stringency we observe likely allows eukaryotes to be responsive to many different environmental bacteria, whilst maintaining tight regulation over important developmental decisions.
Interestingly, we find that certain bacterial genera (including Flavobacteriia, Pseudoalteromonas, and Vibrio) have a high level of influence on the development of diverse marine eukaryotes. It is possible that many eukaryotes may rely on cues from Bacteroidetes and Gamma-proteobacteria because these bacteria flourish in nutrient and carbon rich environments, and can thus serve as proxies for favorable environmental conditions. Indeed, many of the bacteria that regulate eukaryotic development are well equipped for rapidly responding to increasing nutrient availability, boasting an assortment of extracellular enzymes that break down polysaccharides, lipids, and proteins [37]. For example, the polysaccharide degrading abilities of Bacteroidetes and Gammaproteo-bacteria allow these bacteria to utilize algal-derived polysaccharides and promptly proliferate when phytoplankton bloom (often as a result of increased inorganic nutrient levels) [38]. Even more impressive is the ability of some inducing-bacteria (notably Vibrio spp.) to pursue nutrient dense microenvironments through chemotaxis [39]. Thus, diverse eukaryotes may have converged on certain bacteria as indicators of nutrient-rich environments.
Finally, it is enticing to consider how environmental bacteria might benefit from these interactions. Many of the bacteria that induce eukaryotic developmental transitions also frequently associate with eukaryotes, for example by accumulating on surfaces of macroalgae and invertebrates. Because these bacteria produce exoenzymes that help them utilize plant and animal-derived molecules for nutrition [37], it is possible that inducing eukaryotic development allows specific bacteria to rapidly colonize valuable ‘real estate.’
Conclusion
Although the influences of environmental bacteria on the development of marine eukaryotes has been observed for decades, we are just beginning to gain a molecular understanding of these interactions. With the help of model systems and straightforward bioassays, it is becoming clear that environmental bacteria produce structurally diverse cues that govern eukaryotic development with a high degree of molecular specificity. Nonetheless, even seemingly simple bacterial-eukaryotic interactions can be challenging to characterize when we rely solely on activity-guided fractionation. Furthermore, we are finding that most interactions with bacteria are complex, and may rely upon multiple molecular cues produced by one or more bacterial species. To understand these intricate interactions, future studies should likely combine genetic and activity-guided approaches in the study of environmen-tally-relevant pairs or communities of eukaryotes and bacteria.
Importantly, many of the environmental bacteria that regulate eukaryotic development in marine ecosystems interact with eukaryotes in other pathogenic and mutualistic contexts. For example, the bacterium Vibrio fischeri forms a symbiosis with and triggers morphogenesis in the Hawaiian bobtail squid (Euprymna scolopes), while other members of the Vibrionaceae are common animal enteric commensals, pathogens, and mutualists [40]. In addition, Bacteroidetes bacteria closely related to Algoriphagus are abundant mammalian gut commensal bacteria that are important for proper intestinal development and homeostasis [41]. Therefore, understanding how environmental bacteria shape the development of marine eukaryotes may also provide insight into broadly applicable mechanisms of bacterial-eukaryotic interactions.
Acknowledgements
We thank D. Booth, E. Ireland, B. Larson, and T. Linden for providing feedback on the review prior to submission. Research in the King laboratory is supported by the Howard Hughes Medical Institute, the National Institutes of Health (R01GM099533), and the Gordon and Betty Moore Foundation.
References
- 1.Mcfall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF et al. : Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 2013, 110:3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A: The importance of the microbiome of the plant holobiont. New Phytol 2015, 206:1196–1206. [DOI] [PubMed] [Google Scholar]
- 3.Badri DV, Weir TL, van der Lelie D, Vivanco JM: Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol 2009, 20:642–650. [DOI] [PubMed] [Google Scholar]
- 4.McFall-Ngai MJ, Ruby EG: Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 1991, 254:1491–1494. [DOI] [PubMed] [Google Scholar]
- 5.Sommer F, Bäckhed F: The gut microbiota — masters of host development and physiology. Nat Rev Microbiol 2013, 11:227–238. [DOI] [PubMed] [Google Scholar]
- 6.Mcfall-Ngai M: Divining the essence of symbiosis: insights from the squid-Vibrio model. PLoS Biol 2014, 12:e1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster JS, Apicella MA, McFall-Ngai MJ: Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol 2000, 226:242–254. [DOI] [PubMed] [Google Scholar]
- 8.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ: Microbial factor-mediated development in a host-bacterial mutualism. Science 2004, 306:1186–1188. [DOI] [PubMed] [Google Scholar]
- 9.Aschtgen MS, Lynch JB, Koch E, Schwartzman J, Mcfall-Ngai M, Ruby E: Rotation of Vibrio fischeri flagella produces outer membrane vesicles that induce host development. J Bacteriol 2016, 198:2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leadbeater BSC: The Choanoflagellates: Evolution, Ecology, and Biology. Cambridge University Press; 2015. [Google Scholar]
- 11.James-Clark H: On the Spongiae ciliatae as Infusoria flagellata; or, observations on the structure, animality and relationship of Leucosolenia botryoides Bowerbank. Ann Mag Nat His 1868, 1:133–142. [Google Scholar]
- 12.Brunet T, King N: The origin of animal multicellularity and cell differentiation. Dev Cell 2017, 43:124–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairclough SR, Dayel MJ, King N: Multicellular development in a choanoflagellate. Curr Biol 2010, 20:R875–R876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N: Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol 2011, 357:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N: A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. Elife 2012, 1:e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.••.Woznica A, Cantley AM, Beemelmanns C, Freinkman E, Clardy J,· King N: Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc Natl Acad Sci USA 2016, 113:7894–7899. This study describes how rosette development in the choanoflagellate S. rosetta is regulated by a single bacterium that produces a network of activating, enhancing, and inhibitory lipid cues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantley AM, Woznica A, Beemelmanns C, King N, Clardy J: Isolation and synthesis of a bacterially produced inhibitor of rosette development in choanoflagellates. J Am Chem Soc 2016, 138:4326–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin TC, King N: Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Curr Biol 2013, 23:2176–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr M, Leadbeater BSC, Baldauf SL: Conserved meiotic genes point to sex in the choanoflagellates. J Eukaryot Microbiol 2010, 57:56–62. [DOI] [PubMed] [Google Scholar]
- 20.Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ et al. : Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol 2013, 14:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin TC, Greaney AJ, Wetzel L, King N: The Rosetteless gene controls development in the choanoflagellate S. rosetta. Elife 2014:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.••.Woznica A, Gerdt JP, Hulett RE, Clardy J, King N: Mating in the ·closest living relatives of animals is induced by a bacterial chondroitinase. Cell 2017, 170:1175–1183.e11. This study demonstrates that sexual reproduction in the choanoflagellate S. rosetta is induced by a chondroitin-degrading enzyme produced by Vibrio fischeri bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulp A, Kuehn MJ: Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 2010, 64:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch JB, Alegado RA: Spheres of hope, packets of doom: the good and bad of outer membrane vesicles in interspecies and ecological dynamics. J Bacteriol 2017, 199:e00012–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones BW, Maruyama A, Ouverney CC, Nishiguchi MK: Spatial and temporal distribution of the Vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb Ecol 2007, 54:314–323. [DOI] [PubMed] [Google Scholar]
- 26.Wichard T: Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Front Plant Sci 2015, 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S: Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J 2011, 5:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh RP, Reddy CRK: Seaweed-microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol Ecol 2014, 88:213–230. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo Y, Suzuki M, Kasai H, Shizuri Y, Harayama S: Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ Microbiol 2003, 5:25–35. [DOI] [PubMed] [Google Scholar]
- 30.••.Matsuo Y, Imagawa H, Nishizawa M, Shizuri Y: Isolation of an··algal morphogenesis inducer from a marine bacterium. Science 2005, 307:1598 In this paper, the authors isolate Thallusin, a potent inducer of thallus differentiation in Monostroma oxyspermum from an algal-associated environmental bacterium identified in a previous study [29]. [DOI] [PubMed] [Google Scholar]
- 31.Nishizawa M, lyenaga T, Kurisaki T, Yamamoto H, Sharfuddin M, Namba K, Imagawa H, Shizuri Y, Matsuo Y: Total synthesis and morphogenesis-inducing activity of (±)-thallusin and its analogues. Tetrahedron Lett 2007, 48:4229–4233. [Google Scholar]
- 32.•.Spoerner M, Wichard T, Bachhuber T, Stratmann J, Oertel W: Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factors. J Phycol 2012, 48:1433–1447. In this study, the authors determine that the combined activity of two regulatory factors, each factor produced by a different Ulva-associated bacterium, is required for normal cell growth and thallus development in Ulva mutabilis. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Hadfield MG: Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Marine Ecol Progress 2003, 260:161–172. [Google Scholar]
- 34.••.Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL,· Steinberg PD, Harder T : Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a pseudoalteromonas bacterium. PLoS ONE 2011, 6: e19082–e19088. In this study, the authors screen hundreds of environmental bacteria and identify three closely-related strains of Pseudomonas that are sufficient to induce metamorphosis in Acropora millepora. The authors then use activity-guided fractionation to isolate the metamorphic cue tetrabromopyrrole. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.••.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ,· Newman DK: Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 2014, 343:529–533. Using genetic techniques in the bacterium Pseudoalteromonas luteovio-lacea, this study demonstrates that P. luteoviolacea produces contractile arrays of phage tail-like structures that induce metamorphosis in the tubeworm Hydroides elegans, providing mechanistic insight into how bacterial biofilms can trigger larval metamorphosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beemelmanns C, Woznica A, Alegado RA, Cantley AM, King N, Clardy J: Synthesis of the rosette-inducing factor RIF-1 and analogs. J Am Chem Soc 2014, 136:10210–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stal LJ, Cretoiu MS (Eds): The Marine Microbiome. Springer; 2016. [Google Scholar]
- 38.Teeling H, Fuchs BM, Bennke CM, Kruger K, Chafee M, Kappelmann L, Reintjes G, Waldmann J, Quast C, Glockner FO et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. Elife 2016, 5:e11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stocker R: Marine microbes see a sea of gradients. Science 2012, 338:628–633. [DOI] [PubMed] [Google Scholar]
- 40.Thompson FL, Austin B, Swings J: The Biology of Vibrios. ASM Press; 2006. [Google Scholar]
- 41.Thomas F, Hehemann J-H, Rebuffet E, Czjzek M, Michel G: Environmental and gut bacteroidetes: the food connection. Front Microbiol 2011, 2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grueneberg J, Engelen AH, Costa R, Wichard T: Macroalgal morphogenesis induced by waterborne compounds and bacteria in coastal seawater. PLoS ONE 2016, 11:e0146307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall K, Joint I, Callow ME, Callow JA: Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb Ecol 2006, 52:302–310. [DOI] [PubMed] [Google Scholar]
- 44.Singh RP, Mantri VA, Reddy CRK, Jha B: Isolation of seaweed-associated bacteria and their morphogenesis-inducing capability in axenic cultures of the green alga Ulva fasciata. Aquat Biol 2011, 12:13–21. [Google Scholar]
- 45.Joint I, Tait K, Callow ME, Callow JA, Milton D, Williams P, Cámara M: Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 2002, 298:1207. [DOI] [PubMed] [Google Scholar]
- 46.Whalan S, Webster NS: Sponge larval settlement cues: the role of microbial biofilms in a warming ocean. Sci Rep 2014, 4: srep04072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neumann R: Bacterial induction of settlement and metamorphosis in the planula larvae of Cassiopea andromeda. Mar Ecol Prog Ser 1979, 1:21–28. [Google Scholar]
- 48.Schmahl G: Induction of stolon settlement in the scyphopolyps of Aurelia aurita by glycolipids of marine bacteria. Helgolthder wiss Meeresuntersuchungen 1985, 39:117–127. [Google Scholar]
- 49.Aschtgen MS, Wetzel K, Goldman W, Mcfall-Ngai M, Ruby E: Vibrio fischeri-derived outer membrane vesicles trigger host development. Cell Microbiol 2016, 18:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitt WK, Coon SL, Walch M, Weiner RM, Colwell RR, Bonar DB: Settlement behavior and metamorphosis of oyster larvae (Crassostrea gigas) in response to bacterial supernatants. Mar Biol 1990, 106:389–394. [Google Scholar]
- 51.•.Freckelton ML, Nedved BT, Hadfield MG: Induction of invertebrate larval settlement; different bacteria, different mechanisms? Sci Rep 2017, 7:42557 In this study, the authors find that several bacterial species capable of inducing Hydroides elegans larval metamorphosis lack the ability to produce phage tail-like structures (see [35··]), indicating that H. elegans can respond to diverse metamorphic cues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huggett MJ, Williamson JE, De Nys R, Kjelleberg S, Steinberg PD: Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 2006, 149:604–619. [DOI] [PubMed] [Google Scholar]
- 53.Szewzyk U, Holmström C, Wrangstadh M: Relevance of the exopolysaccharide of marine Pseudomonas sp. strain S9 for the attachment of Ciona intestinalis larvae. Ecol Progress Ser 1991. 10.2307/24825861. [DOI] [Google Scholar]


