FIG. 2.
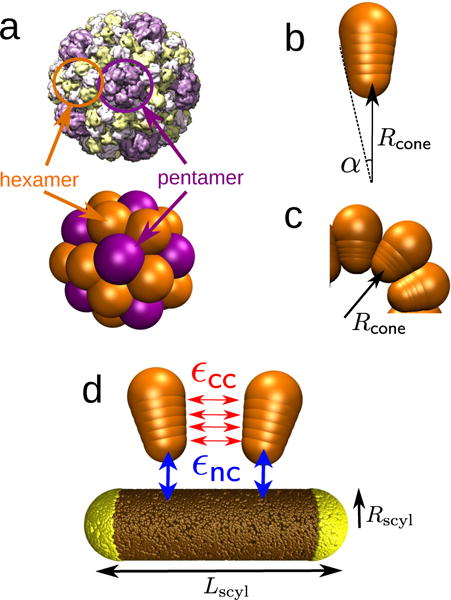
(a) Brome mosaic virus (BMV) capsid structure obtained from cyoEM (PDB ID: 3J7M) [79], compared with a capsid assembled from conical subunits in the absence of a nanoparticle. Each conical subunit represents a capsomer, and both pentamers (five neighbors, in purple) and hexamers (six neighbors, in orange) can be identified, showing icosahedral organization consistent with T=3 symmetry [22, 80]. (b,c) The cone angle α (b) sets the capsid spontaneous curvature radius Rcone (c), defined as the radial position of the capsomer bottom bead in the lowest energy capsid. (d) Capsomer model for Rcone = 9.5nm. Each of four interior beads (indices n = 1 … 4) interacts through a Morse potential of depth εcc with its counterparts in neighboring capsomers (red). The bottom capsomer bead experiences attractive interactions of strength εnc with the nanoparticle beads (blue). The equilibrium distance of the Morse potential between nanoparticle and capsomer beads, , must be considered in the template curvature. The effective template radius is then given by .
