Abstract
BACKGROUND: Signet-ring cell carcinoma (SRCC) is a very rare subtype of colorectal adenocarcinoma (COAD) with a poor clinical prognosis. Although understanding key mechanisms of tumor progression in SRCCs is critical for precise treatment, a comprehensive view of genomic alterations is lacking. MATERIALS AND METHODS: We performed whole-exome sequencing of tumors and matched normal blood as well as RNA sequencing of tumors and matched normal colonic tissues from five patients with SRCC. RESULTS: We identified major somatic alterations and characterized transcriptional changes at the gene and pathway level. Based on high-throughput sequencing, the pattern of mutations and copy number variations was overall similar to that of COAD. Transcriptome analysis revealed that major transcription factors, such as SRF, HNF4A, ZEB1, and RUNX1, with potential regulatory roles in key pathways, including focal adhesion, the PI3K-Akt signaling pathway, and the MAPK signaling pathway, may play a role in the tumorigenesis of SRCC. Furthermore, significantly upregulated genes in SRCCs were enriched for epithelial-mesenchymal transition genes, and accumulation of mucin in intracytoplasm was associated with the overexpression of MUC2. CONCLUSION: The results indicate that the molecular basis of colorectal SRCC exhibits key differences from that of consensus COAD. Our findings clarify important genetic features of particular abnormalities in SRCCs.
Introduction
Colorectal cancer is the third most common malignancy and a leading cause of cancer-related deaths worldwide [1], [2]. Colorectal cancer represents a group of histologically heterogeneous tumors, and its histological subtype is determined by the major component of cancer cells [3], [4], [5]. There are several histological subtypes in colorectal cancer; adenocarcinoma is the most common type, and other subtypes include mucinous carcinoma, signet-ring cell carcinoma (SRCC), squamous cell carcinoma, and undifferentiated carcinoma [2], [3], [4]. The identification of the histological subtype is important for cancer patient management because it plays an important role in determining tumor biology and aggressiveness. In colorectal cancer, histological subtype is considered an important prognostic factor.
SRCC is a rare subtype of colorectal cancer, accounting for approximately 1% of patients with colorectal cancer [5], [6]. Signet-ring cell is characterized by intracytoplasmic mucin that displaces the nucleus to one side. SRCC is defined as a carcinoma with a signet-ring cell component of greater than 50% [4], [6]. Previous studies have reported that colorectal SRCC is associated with young age, advanced tumor stage, high rates of metastasis, and poor prognosis [5], [7]. These aggressive behaviors are associated with specific molecular features such as microsatellite instability, a high frequency of CpG island methylator phenotypes, and frequent BRAF and KRAS mutations [3], [4]. Despite the poor prognosis, limited studies have characterized colorectal SRCC owing to the low incidence. Thus, clinicopathological features are not well understood, and few studies have compared SRCC with typical adenocarcinoma of colorectal cancer, especially at the molecular level. In addition, the genomic characteristics of SRCC have been only partially defined by low-throughput molecular studies so that comprehensive genomic profiling is needed for better understanding molecular mechanisms of SRCC.
In this study, we conducted a comprehensive analysis of five SRCCs by both whole-exome and RNA sequencing with tumors and matched normal. Whole-exome sequencing (WES) identified DNA aberrations in somatic mutations and copy number alterations for individual SRCCs. The transcriptome profiling also identified differentially expressed genes between tumor and normal samples, their involved pathways, and transcriptional regulators that may play a critical role in SRCCs. Our results revealed that the molecular basis of colorectal SRCC exhibits key differences from that of consensus colorectal adenocarcinoma (COAD).
Materials and Methods
Sample Collection
This study was approved by the Institutional Review Board (IRB) of Samsung Medical Center (IRB approval no. SMC 2013-11-008). Written informed consent was obtained from each patient. The study subjects were five Korean patients diagnosed with the SRCC subtype of colorectal cancer at Samsung Medical Center, Seoul, Korea. Tumor and matched normal tissues were obtained from surgical specimen. Peripheral blood mononuclear cells (PBMC) were obtained from each patient. PBMC and normal mucosa were used for genomic and transcriptomic analysis as a control, respectively. Samples were snap-frozen and stored in liquid nitrogen until use. Histology, clinical stage, and analyzed lesions are summarized in Table 1. Hematoxylin and eosin staining results for patient tumor samples are shown in Supplementary Figure 1.
Table 1.
Patient Characteristics of Five SRCCs
| Characteristics | SRCC1 | SRCC2 | SRCC3 | SRCC4 | SRCC5 |
|---|---|---|---|---|---|
| Age | 46 | 40 | 38 | 60 | 29 |
| Gender | M | M | M | M | F |
| Tumor location | Descending colon | Rectum | Ascending colon | Rectum | Rectosigmoid junctions |
| MSI status | MSS | MSS | MSS | MSS | MSS |
| TNM stage | IVB | IVA | IVB | IVB | IVA |
| Metastasis | Yes | Yes | Yes | Yes | Yes |
| Lymphatic invasion | Positive | Positive | Negative | Positive | Positive |
| Perineural invasion | Negative | Negative | Positive | Negative | Negative |
| Vascular invasion | Negative | Negative | Positive | Positive | Positive |
TCGA Colorectal Cancer Data Sets
We downloaded the 629 clinical, 489 raw mutation annotation, 623 mRNASeq, and 616 SNP6 copy number of COAD from Broad GDAC firehose (http://gdac.broadinstitute.org, version: 2016.01.28). Based on clinical information, all COADs are adenocarcinoma or mucinous carcinoma. Therefore, we used TCGA COAD data sets as a reference in this study. We choose 32 of 623 samples for differentially expressed gene analysis because those had matched normal-tumor pairs. Lists of TCGA samples and its clinicopathological features are summarized in Supplementary Table 3.
Isolation of Genomic DNA and RNA
Genomic DNA and RNA in tissues were purified using the AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA). Genomic DNA from peripheral blood was extracted using the QIAamp DNA Blood Mini Kit (Qiagen). Genomic DNA concentration and purity were measured using a NanoDrop 8000 UV-Vis Spectrometer (Thermo Scientific Inc., Wilmington, DE) and a Qubit 2.0 Fluorometer (Life Technologies Inc., Grand Island, NY). To estimate DNA degradation, median DNA size and ΔCt values were measured using a 2200 TapeStation Instrument (Agilent Technologies, Santa Clara, CA) and real-time PCR (Agilent Technologies), respectively. For RNA, concentration and purity were measured using the NanoDrop and Bioanalyzer (Agilent Technologies).
Whole-Exome Sequencing
Genomic DNA (1 μg) from each sample was sheared using the Covaris S220 (Covaris, Woburn, MA) and used to construct a library using SureSelect XT Human All Exon V5 and a SureSelect XT Reagent Kit, HSQ (Agilent Technologies) according to the manufacturer’s protocol. This kit is designed to enrich 335,756 exons of 21,058 genes, covering ~71 Mb of the human genome. After enriched exome libraries were multiplexed, the libraries were sequenced using a HiSeq 2500 sequencing platform (Illumina, San Diego, CA). Briefly, a paired-end DNA sequencing library was prepared by gDNA shearing, end-repair, A-tailing, paired-end adaptor ligation, and amplification. After hybridization of the library with bait sequences for 16 hours, the captured library was purified and amplified with an index barcode tag, and library quality and quantity were measured. Sequencing of the exome library was carried out using the 100-bp paired-end mode of the TruSeq Rapid PE Cluster Kit and TruSeq Rapid SBS Kit (Illumina). Sequencing data and results are summarized in Supplementary Table 1.
Exome-seq Data Analysis
Sequencing reads were aligned to the UCSC hg19 reference genome (downloaded from http://genome.ucsc.edu), using Burrows-Wheeler Aligner [8] version 0.7.5a, with default settings. PCR duplicates were marked using Picard-tools-1.93 (http://picard.sourceforge.net/), data cleanup was performed with GATK, and variants were identified with GATK-3.5 [9]. Then, point mutations were identified using MuTect [10] and VarScan2 [11] with paired samples. An in-house Perl script and ANNOVAR [12] were used to annotate variants. Tumor purity estimation based on WES was performed with THetA [13] (Supplementary Figure 2). Somatic copy number alterations were identified using CNVkit [14], and the resulting ratio was adjusted according to tumor purity.
RNA Sequencing
Library construction for RNA sequencing was performed using a Truseq RNA Sample Preparation v2 Kit (Illumina). Isolated total RNA was used in a reverse transcription reaction with poly (dT) primers, using SuperScript II Reverse Transcriptase (Invitrogen/Life Technologies) according to the manufacturer’s protocol. Briefly, an RNA sequencing library was prepared by cDNA amplification, end-repair, 3′ end adenylation, adapter ligation, and amplification. Library quality and quantity were measured using the Bioanalyzer and Qubit. Sequencing of the RNA library was carried out using the 100-bp paired-end mode of the TruSeq Rapid PE Cluster Kit and the TruSeq Rapid SBS Kit (Illumina).
RNA-seq Data Analysis
Reads from the FASTQ files were mapped to the hg19 human reference genome using STAR version 2.5.0a in 2-pass mode [15], and gene quantification was performed using RSEM [16]. Expressed genes were defined as those with a transcripts per million (TPM) value of more than 10 across all samples to reduce the false-positive rate. Differentially expressed genes (DEGs) were identified using the edgeR package version 3.14.0 with a cutoff (|log2 fold-change| > 2 and false discovery rate (FDR) < 0.001) [17]. DEGs were mapped to the pathway using the REACTOME pathway database [18]. Tumor purity based on whole-transcriptome sequencing was performed with ESTIMATE [19] (Supplementary Figure 2). Fusion genes were detected using deFuse [20] (v0.6.1) and JAFFA (v1.06) [21] with default settings. The overlapped fusion transcripts between two prediction tools were considered as candidates.
Prediction of Transcription Regulators
iRegulon (Cytoscape plugin) detects master transcription regulators from a set of DEGs [22]. Regulators were predicted among the set of DEGs using iRegulon. Briefly, Cytoscape networks were created by importing the list of DEGs. The set of nodes (genes) was submitted to iRegulon and analyzed using the following options: 1) motif collection (10-kb region, 9,713 PWMs), 2) track collection (750 ChIP-seq tracks of ENCODE uniform signals), 3) putative regulatory region (10kb centered around TSS), 4) motif rankings database (10-kb region centered around TSS, 7 species), and 5) track of rankings database (10-kb region centered around TSS, ChIP-seq-derived).
Gene Set Enrichment Analysis
A gene set enrichment analysis (GSEA) [23] was conducted to analyze an SRCC-specific upregulated gene set. A GSEA preranked analysis was performed by inputting a list of genes sorted according to fold changes of each TCGA COAD and SRCC.
qPCR Assay
Total RNA was extracted from patient tissue (RNAprep Mini kit, Qiagen), and 500 ng RNA was subjected to reverse transcription using reverse transcription kit (Bioneer). Real-time quantitative PCR amplification was performed with a SYBR Green (ABI) in a real-time system (ABI, USA). Human-specific PCR primers (Bioneer) were used to analyze expression of the following genes: TWIST1, SNAI1, SNAI2, ZEB1, and GAPDH. mRNA levels of specific genes were calculated as ΔΔCt and normalized to GAPDH.
Western Blot
To prepare tissue extracts, tissues were lysed using a protein lysis buffer including a protease inhibitor. Then, 30 to 60 μg of protein extract was incubated with primary antibodies against TWIST1 (ab50887, Abcam), SNAI1 (#3895, Cell Signaling), SNAI2 (ab38551, Abcam), and ZEB1 (sc-25388, Santa Cruz). β-Actin (#3700, Cell Signaling) was used as normalized protein controls in Western blotting.
Results
Somatic Alterations in Colon SRCC
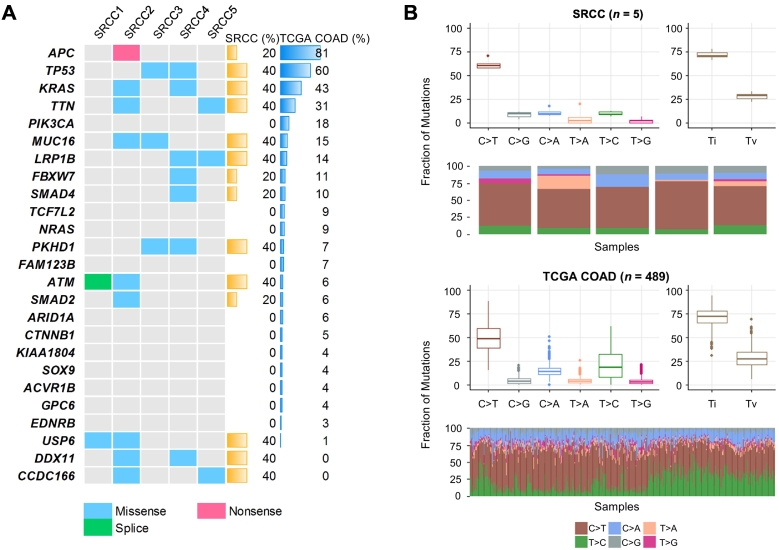
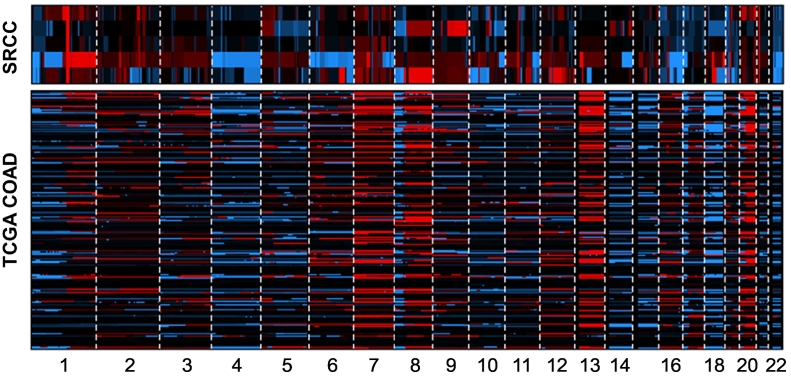
Somatic mutations frequently observed in TCGA nonhypermutated COADs [24] were also identified in SRCCs (Figure 1A, Supplementary Figure 3). Most mutations were commonly found in SRCCs and TCGA COADs. However, APC nonsense mutation was observed at only one case (SRCC2) among five SRCCs (binomial test, P = .006), but there was a high frequency in COADs (81%). Mutations in three genes (USP6, DDX11, and CCDC166) having a very low frequency in COADs (≤1%) were relatively high in SRCCs (Supplementary Figure 4). Next, we performed a sequence context analysis (Figure 1B). Transition/transversion ratios were broadly consistent with those of COADs. A high frequency of C>T transitions is predominantly observed in COADs [25], and similarly, this transition type represented a large proportion of sequence changes in SRCCs. Furthermore, we identified chromosomal alterations in SRCCs. According to previous studies, COADs have recurrent chromosomal alterations, such as gains of 1q, 7p and q, 8p and q, 12q, 13q, 19q, and 20p and q and deletions of 1p, 4q, 5q, 8p, 14q, 15q, 17p and q, 18p and q, 20p, and 22q [24], [26]. These alterations were consistently detected in SRCCs (Figure 2). Taken together, the results of somatic mutation and copy number alteration analyses at the whole-exome level revealed that SRCCs and COADs have overall similar properties at the genomic level, but APC gene mutation was uncommonly detected.
Figure 1.
Somatic mutations and sequence context of SRCC. Notes: (A) Heatmap of somatic mutations (including missense, nonsense, and splicing mutations) detected in SRCCs. Genes are sorted by the mutation frequency observed in TCGA nonhypermutated COADs. (B) Comparison of sequence context between SRCCs and TCGA COADs. The spectrum of substitutions, including six classes, and the Ti/Tv rate are shown. Abbreviations: Ti, transition; Tv, transversion.
Figure 2.
Copy number alterations in both SRCCs and TCGA COADs. Notes: The copy number alterations are plotted across chromosomes (red: amplification, blue: deletion, black: diploid).
In addition, we found four novel fusion transcripts in the two SRCC cases (Supplementary Figure 5, A and B). One of the four fusions was interchromosomal fusion, and the remaining three were intrachromosomal fusions. Three of the four fusions showed the highest expression fold change (tumor vs. normal) of the partner gene in samples with fusion among the five SRCC samples (Supplementary Figure 5C).
Differentially Expressed Genes and Their Transcriptional Regulators
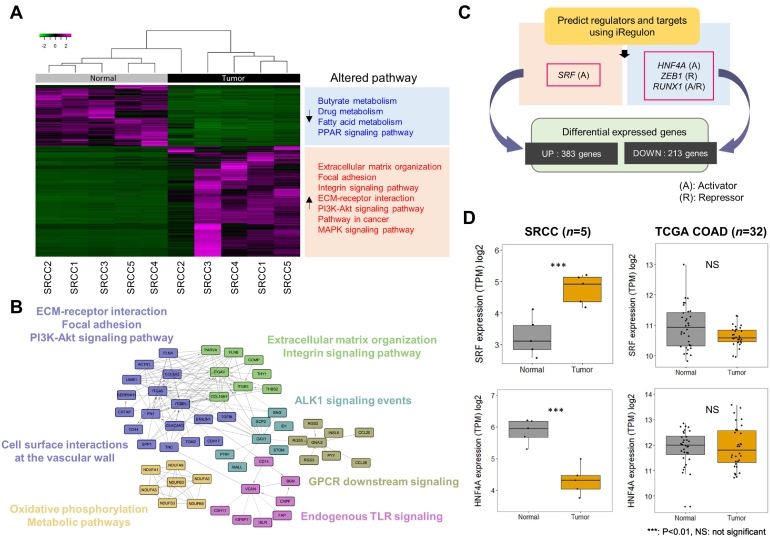
We investigated DEGs between SRCC tumors and matched normal colonic tissues. We detected 596 DEGs (upregulated: 383; downregulated: 213) with |log2 fold-change| > 2 and FDR < 10−3 as criteria and mapped these genes to pathways (Figure 3, A and B). Upregulated genes were included in several pathways, including “extracellular matrix (ECM) organization,” “focal adhesion,” “integrin signaling pathway,” and “ECM-receptor interaction,” and these genes may be related to phenotypic traits of signet-ring cells and metastasis ability [27], [28], [29], [30]. Other upregulated pathways were involved in cell survival and proliferation, such as the “PI3K/AKT and MAPK signaling pathways,” which are well-known signatures of cancer cells [31], [32], [33], [34], [35].
Figure 3.
Differentially expressed genes, pathways, and their transcription factors. Notes: (A) Heatmap of differentially expressed genes in SRCCs. Enriched pathways among up/downregulated genes are also presented. (B) Pathway-based functional interaction network. Genes belonging to same pathway are closely linked to each other. (C) Prediction of transcription factors that regulate differentially expressed genes. (D) Boxplot of the expression levels of SRF and HNF4A. Expression levels in TCGA COADs with tumor and matched normal samples were compared.
To identify the master transcription factors that regulate the DEGs, we conducted an iRegulon analysis (Figure 3C). We found four transcription factors (SRF, HNF4A, ZEB1, and RUNX1) that were significantly enriched in DEGs and compared their expression levels between tumor and normal tissues for each SRCC and TCGA COAD (Figure 3D, Supplementary Figure 6). All four transcription factors exhibited significantly altered gene expression levels (tumor vs. normal), and three of four showed different patterns in comparisons with TCGA COADs. For example, serum response factor (SRF) is a transcription activator that binds to the serum response element in the promoter region of target genes [36]. SRF gene expression was highly elevated in tumors compared with normal tissues in SRCCs but not in TCGA COADs. Hepatocyte nuclear factor 4 alpha (HNF4A) is a nuclear transcription factor that regulates the morphology and function of epithelial cells, and its gene expression was lower in SRCC tumors than normal tissues. These results indicate that altered expression levels of these transcription factors play a role in the tumorigenesis of SRCC.
SRCC-Specific Upregulated Genes
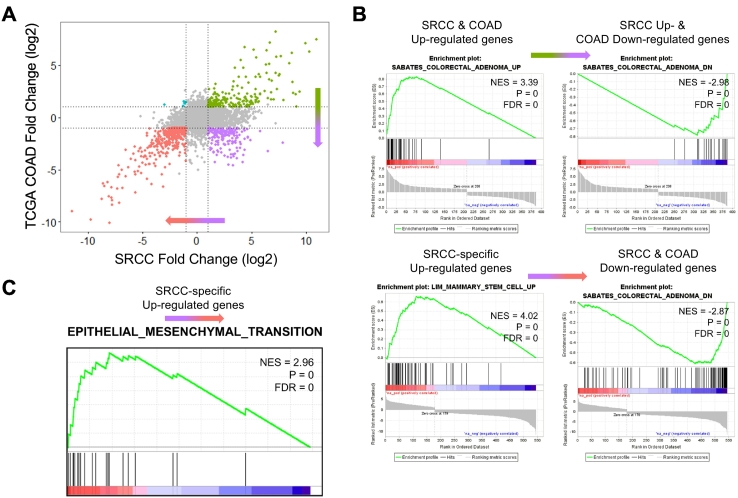
To identify signet-ring cell–specific dysregulated genes, we compared the expression levels between five SRCCs and 32 TCGA COADs (Figure 4A). After the removal of genes that did not exhibit expression differences (P > .01), we found 759 genes (208 up- and 366 downregulated genes in both SRCCs and COADs, 179 upregulated in SRCCs but downregulated in COADs, and 6 downregulated in SRCCs but upregulated in COADs). To identify the functional roles of SRCC-specific upregulated genes (179 genes; Figure 4A, bottom-right), we performed a GSEA. First, we selected the genes that had high fold-change values in SRCCs, which included the upregulated genes of both COADs and SRCCs and SRCC-specific upregulated genes. Then, we ordered genes by the fold change values (Figure 4A, from top-right to bottom-right). As expected, enriched gene sets were “Colorectal Adenoma Up” and “Colorectal Adenoma Down” (Figure 4B upper). Second, we selected and sorted genes that had low fold-change values in COADs. Interestingly, SRCC-specific upregulated genes were enriched in gene sets of “Stem Cell Up Regulation” and “Epithelial-Mesenchymal Transition (EMT)” (Figure 4B lower and Figure 4C). The EMT is a process involved in the loss of polarity in epithelial cells and cell-cell adhesion. The progression to mesenchymal cells is related to the acquired migration ability, invasiveness, and increased production of ECM components [37]. In addition, this EMT process can generate cells with stem cell–like properties [38]. Taken together, our results suggest that SRCCs may acquire potential stem cell–like characteristics via the EMT process.
Figure 4.
SRCC-specific highly expressed genes and enrichment analysis. Notes: (A) Scatter plot of log2-fold changes in expression (tumor vs. normal) for SRCCs and COADs (green: upregulated in both SRCCs and COADs, purple: upregulated in SRCCs but downregulated in COAD, red: downregulated in both SRCCs and COADs; cyan: downregulated in SRCCs but upregulated in COAD). (B-C) Preranked GSEA. Genes were sorted based on log2-fold change.
Signature of Epithelial-Mesenchymal Transition and Mucin Production
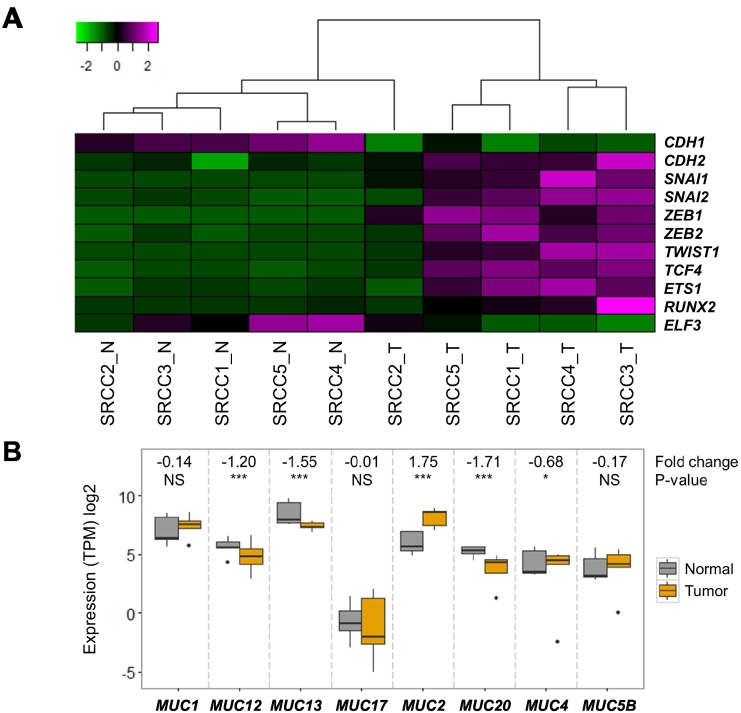
The main characteristics of signet-ring cells are a loss of cell-cell interactions and accumulation of intracellular mucin. Typically, epithelial cells express high levels of E-cadherin (CDH1), whereas mesenchymal cells express N-cadherin (CDH2) [39]. We investigated the expression of genes encoding E and N-cadherin and their transcription factors to estimate the EMT signature in SRCCs (Figure 5A). The expression of E-cadherin was lower in tumor tissues than in normal tissues, whereas N-cadherin expression was higher in tumor tissues than in normal tissues. Gene expression levels of transcription factors that directly or indirectly regulate E-cadherin, such as SNAI1, SNAI2, ZEB1, ZEB2, TWIST1, TCF4, ETS1, and RUNX2, increased during the tumorigenesis of signet-ring cells. In addition, ELF3 is a negative regulator of SNAI1, ZEB, and TWIST1, which exhibit decreased expression in SRCC tumors [40]. These results indicate that SRCCs undergo the EMT process, which is the main cause of the loss of cell-cell interactions.
Figure 5.
Expression of epithelial-mesenchymal transition markers and MUC2 overexpression. Notes: (A) Heatmap of the expression levels of epithelial-mesenchymal transition markers. (B) Boxplot of the expression levels of mucin-producing genes.
To determine which mucin-producing genes are activated in SRCCs, we compared expression levels in tumor vs. normal samples of human mucin genes (Figure 5B). Nonexpressed mucin genes are not shown here. Among mucin genes, MUC2 was more highly expressed (3.7-fold) in tumor than in normal tissues, but other genes were either decreased or unchanged. In COADs, the expression levels of all mucin genes, including MUC2, decreased (Supplementary Figure 7). These results indicate that mucin accumulation in signet-ring cells is caused by the abnormal expression of MUC2 [41], [42], [43].
Discussion
In this study, we first characterized somatic alterations on the genome-wide scale for colorectal SRCC by whole-exome and RNA sequencing. Previous SRCC studies are focused only few cancer gene mutations and expressions or case reports. However, this work provides genomic and transcriptomic alterations based on massive parallel sequencing simultaneously. As compared with those for SRCCs and TCGA COADs, we demonstrated interesting findings that have not been previously reported about SRCCs. Mutations in the APC gene are the initial genetic alterations in colorectal tumorigenesis [44]. However, the low APC mutation rate in SRCCs indicates that they undergo a developmental process different from that of COAD. We also validated of low APC mutation rate (17%) in additional 6 SRCC cases (Supplementary Figure 9A). A previous study reported a distinct pattern of KRAS gene mutations in SRCCs; specifically, there is a lower mutation frequency at codon 12 (2 of 16) but a higher mutation frequency at codon 61 (4 of 16) as compared with those of non-SRCCs [45]. In our study, two samples had KRAS mutations at codon 12 (SRCC2: G12C, SRCC4: G12D), and mutation at codon 61 was not detected. A BRAF mutation was not detected in our SRCCs (Figure 1A, Supplementary Figure 9A). This might be explained by their microsatellite stable (MSS) status. Previous studies have reported a high frequency of BRAF (V600E) mutations in SRCCs, including high-level microsatellite instability (MSI-H) [46], [47], [48]. In TCGA COAD with hypermutated tumors, the mutation frequency in BRAF (V600E) is approximately 41% [24], but this frequency is much lower in nonhypermutated tumors. We have also validated the gene mutations through the Sanger sequencing and both whole-exome and RNA sequencing (Supplementary Figure 3).
Recurrent fusion genes in colorectal cancer [49] were not detected in SRCC samples, but we found several novel fusion genes (Supplementary Figure 5). However, the functional implications of these fusion genes are unknown. We found SRCC-specific expression changes for transcription factors such as HNF4A, ZEB1, and RUNX1 that may regulate target genes in SRCCs. These target genes are involved in butyrate and fatty acid metabolism, and PPAR signaling pathway. Short-chain fatty acids, which include butyrate, are produced in the human colon by bacterial fermentation and suppressor of colorectal cancer [50], [51], [52]. In addition, peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors that are bound to oxidized fatty acids and regulate the differentiation of cells [53], [54]. This means that butyrate and fatty acid metabolism, and PPAR signaling pathway are related to each other. In SRCCs, butyrate and fatty acid metabolism, and PPAR signaling pathways were all downregulated. Thus, our results support the previous reports about tumor suppressive role of butyrate and fatty acid metabolism, and PPAR signaling pathway in colorectal cancer.
Finally, we examined the relationship between clinicopathological features and genetic abnormalities in SRCCs. Highly upregulated genes were enriched in the EMT process. We selected four of eight transcription factors that regulate highly upregulated genes in SRCCs to measure the mRNA and protein expression levels using qPCR and Western blotting, respectively (Supplementary Figure 8, A and B). Although the mRNA expressions of qPCR are similar to those of RNA-seq, protein expressions are different from those of mRNA expressions (Supplementary Table 2). There may be several reasons for discrepancy between mRNA and protein expressions such as posttranscriptional mechanisms, difference of in vivo proteins half-lives, and experimental error in measuring the amount of both mRNA and protein [55]. In addition, MUC2 overexpression is considered as a major cause of the accumulation of mucin in signet-ring cells. Four transcription factors (SRF, HNF4A, ZEB1, and RUNX1) and MUC2 expressions were also validated using public expression data of SRCCs (GSE79793 [56]), and those gene expressions were corresponded to our study except for ZEB1 (Supplementary Figure 9B). Our results provide important insight to the understanding of molecular alterations in SRCCs.
The following are the supplementary data related to this article.
Sequencing data and results of both WES and RNA sequencing.
Summary of mRNA (RNA-seq and qPCR) and protein (Western blotting) expression change of selected four genes in SRCCs.
List of TCGA samples which were used in this study and its clinicopathological features.
Hematoxylin and eosin staining for patient tumor samples. Representative microscopic photographs of SRCCs, which were composed of almost signet-ring cells.
Tumor purity based on whole-exome and RNA sequencing.
Sanger sequencing and IGV snapshot of detected mutations in SRCC cases. Mutations are validated using Sanger sequencing and IGV of both whole-exome and RNA sequencing.
Novel recurrent mutations of USP6, DDX11, and CCDC166 in SRCCs.
Novel fusion genes in two SRCC cases. (A-B) Putative fusion genes of SRCC2 and SRCC4 respectively. (C) Gene expression of each fusion partner. The x-axis is 5′ partner and the y-axis is 3′ partner. Black dot is a sample with fusion, and the gray dots are samples without fusion.
Expression levels of ZEB1 and RUNX1 in both SRCCs and TCGA COADs. Expression levels of TCGA COADs with tumor and matched normal samples (n = 32) were compared to SRCCs.
Expression levels of mucin produce genes in TCGA COADs.
Validation of mRNA and protein expressions using qPCR and Western blotting. (A) qPCR assay results for TWIST1, SNAI1, SNA2, and ZEB1. (B) Western blot results for TWIST1, SNAI1, SNA2, and ZEB1.
External validation of four transcription factors (SRF, HNF4A, ZEB1, and RUNX1) and MUC2 expression.
Footnotes
Acknowledgements: This research was supported by the Samsung Medical Center and Health Technology R&D Project, the Ministry of Health & Welfare (grant number: HI13C2096 to W.-Y.P.), by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (grant number: NRF-2017M3A9A7050803 to W.-Y.P.), and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C3418 and HI15C1593 to Y.B.C.). The biospecimens for this study were provided by Samsung Medical Center Biobank. All sequencing was performed by Samsung Genome Institute.
Disclosure: The author reports no conflicts of interest in this work.
Contributor Information
Je-Gun Joung, Email: jegun.joung@samsung.com.
Woong-Yang Park, Email: woongyang@skku.edu.
Yong Beom Cho, Email: yongbeom.cho@samsung.com.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Sui X, Xu Y, Yang J, Fang Y, Lou H, Han W, Zhang M, Chen W, Wang K, Li D. Use of metformin alone is not associated with survival outcomes of colorectal cancer cell but AMPK activator AICAR sensitizes anticancer effect of 5-fluorouracil through AMPK activation. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nitsche U, Zimmermann A, Spath C, Muller T, Maak M, Schuster T, Slotta-Huspenina J, Kaser SA, Michalski CW, Janssen KP. Mucinous and signet-ring cell colorectal cancers differ from classical adenocarcinomas in tumor biology and prognosis. Ann Surg. 2013;258(5):775–782. doi: 10.1097/SLA.0b013e3182a69f7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inamura K, Yamauchi M, Nishihara R, Kim SA, Mima K, Sukawa Y, Li T, Yasunari M, Zhang X, Wu K. Prognostic significance and molecular features of signet-ring cell and mucinous components in colorectal carcinoma. Ann Surg Oncol. 2015;22(4):1226–1235. doi: 10.1245/s10434-014-4159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugen N, Verhoeven RH, Lemmens VE, van Aart CJ, Elferink MA, Radema SA, Nagtegaal ID, de Wilt JH. Colorectal signet-ring cell carcinoma: benefit from adjuvant chemotherapy but a poor prognostic factor. Int J Cancer. 2015;136(2):333–339. doi: 10.1002/ijc.28981. [DOI] [PubMed] [Google Scholar]
- 6.Thota R, Fang X, Subbiah S. Clinicopathological features and survival outcomes of primary signet ring cell and mucinous adenocarcinoma of colon: retrospective analysis of VACCR database. J Gastrointest Oncol. 2014;5(1):18–24. doi: 10.3978/j.issn.2078-6891.2013.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung CO, Seo JW, Kim KM, Do IG, Kim SW, Park CK. Clinical significance of signet-ring cells in colorectal mucinous adenocarcinoma. Mod Pathol. 2008;21(12):1533–1541. doi: 10.1038/modpathol.2008.170. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oesper L, Mahmoody A, Raphael BJ. THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol. 2013;14(7):R80. doi: 10.1186/gb-2013-14-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4) doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7(5):e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson NM, Majewski IJ, Oshlack A. JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7(1) doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10(7):e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Wang X, Wu F, Huang R, Xue F, Liang G, Tao M, Cai P, Huang Y. Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28(1-2):35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 30.Kornberg LJ. Focal adhesion kinase and its potential involvement in tumor invasion and metastasis. Head Neck. 1998;20(8):745–752. doi: 10.1002/(sici)1097-0347(199812)20:8<745::aid-hed14>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 31.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, Weiss HL, Evers BM. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210(5):767–776. doi: 10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015;1855(1):104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 35.Setia S, Nehru B, Sanyal SN. Upregulation of MAPK/Erk and PI3K/Akt pathways in ulcerative colitis-associated colon cancer. Biomed Pharmacother. 2014;68(8):1023–1029. doi: 10.1016/j.biopha.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229(1):1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 37.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 41.Borger ME, Gosens MJ, Jeuken JW, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID. Signet ring cell differentiation in mucinous colorectal carcinoma. J Pathol. 2007;212(3):278–286. doi: 10.1002/path.2181. [DOI] [PubMed] [Google Scholar]
- 42.Bu XD, Li N, Tian XQ, Li L, Wang JS, Yu XJ, Huang PL. Altered expression of MUC2 and MUC5AC in progression of colorectal carcinoma. World J Gastroenterol. 2010;16(32):4089–4094. doi: 10.3748/wjg.v16.i32.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh MD, Clendenning M, Williamson E, Pearson SA, Walters RJ, Nagler B, Packenas D, Win AK, Hopper JL, Jenkins MA. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26(12):1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 44.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 45.Wistuba II, Behrens C., Albores-Saavedra J., Delgado R., Lopez F., Gazdar AF. Distinct K-ras mutation pattern characterizes signet ring cell colorectal carcinoma. Clin Cancer Res. 2003;9(10 Pt 1):3615–3619. [PubMed] [Google Scholar]
- 46.Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19(1):59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 47.Kakar S, Deng G, Smyrk TC, Cun L, Sahai V, Kim YS. Loss of heterozygosity, aberrant methylation, BRAF mutation and KRAS mutation in colorectal signet ring cell carcinoma. Mod Pathol. 2012;25(7):1040–1047. doi: 10.1038/modpathol.2012.44. [DOI] [PubMed] [Google Scholar]
- 48.Kakar S, Smyrk TC. Signet ring cell carcinoma of the colorectum: correlations between microsatellite instability, clinicopathologic features and survival. Mod Pathol. 2005;18(2):244–249. doi: 10.1038/modpathol.3800298. [DOI] [PubMed] [Google Scholar]
- 49.Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132(5):1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 51.Sengupta S, Muir JG, Gibson PR. Does butyrate protect from colorectal cancer? J Gastroenterol Hepatol. 2006;21(1 Pt 2):209–218. doi: 10.1111/j.1440-1746.2006.04213.x. [DOI] [PubMed] [Google Scholar]
- 52.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 53.Jackson L, Wahli W, Michalik L, Watson SA, Morris T, Anderton K, Bell DR, Smith JA, Hawkey CJ, Bennett AJ. Potential role for peroxisome proliferator activated receptor (PPAR) in preventing colon cancer. Gut. 2003;52(9):1317–1322. doi: 10.1136/gut.52.9.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogino S, Shima K, Baba Y, Nosho K, Irahara N, Kure S, Chen L, Toyoda S, Kirkner GJ, Wang YL. Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology. 2009;136(4):1242–1250. doi: 10.1053/j.gastro.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9) doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvi MA, Loughrey MB, Dunne P, McQuaid S, Turkington R, Fuchs MA, McGready C, Bingham V, Pang B, Moore W. Molecular profiling of signet ring cell colorectal cancer provides a strong rationale for genomic targeted and immune checkpoint inhibitor therapies. Br J Cancer. 2017;117(2):203–209. doi: 10.1038/bjc.2017.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing data and results of both WES and RNA sequencing.
Summary of mRNA (RNA-seq and qPCR) and protein (Western blotting) expression change of selected four genes in SRCCs.
List of TCGA samples which were used in this study and its clinicopathological features.
Hematoxylin and eosin staining for patient tumor samples. Representative microscopic photographs of SRCCs, which were composed of almost signet-ring cells.
Tumor purity based on whole-exome and RNA sequencing.
Sanger sequencing and IGV snapshot of detected mutations in SRCC cases. Mutations are validated using Sanger sequencing and IGV of both whole-exome and RNA sequencing.
Novel recurrent mutations of USP6, DDX11, and CCDC166 in SRCCs.
Novel fusion genes in two SRCC cases. (A-B) Putative fusion genes of SRCC2 and SRCC4 respectively. (C) Gene expression of each fusion partner. The x-axis is 5′ partner and the y-axis is 3′ partner. Black dot is a sample with fusion, and the gray dots are samples without fusion.
Expression levels of ZEB1 and RUNX1 in both SRCCs and TCGA COADs. Expression levels of TCGA COADs with tumor and matched normal samples (n = 32) were compared to SRCCs.
Expression levels of mucin produce genes in TCGA COADs.
Validation of mRNA and protein expressions using qPCR and Western blotting. (A) qPCR assay results for TWIST1, SNAI1, SNA2, and ZEB1. (B) Western blot results for TWIST1, SNAI1, SNA2, and ZEB1.
External validation of four transcription factors (SRF, HNF4A, ZEB1, and RUNX1) and MUC2 expression.