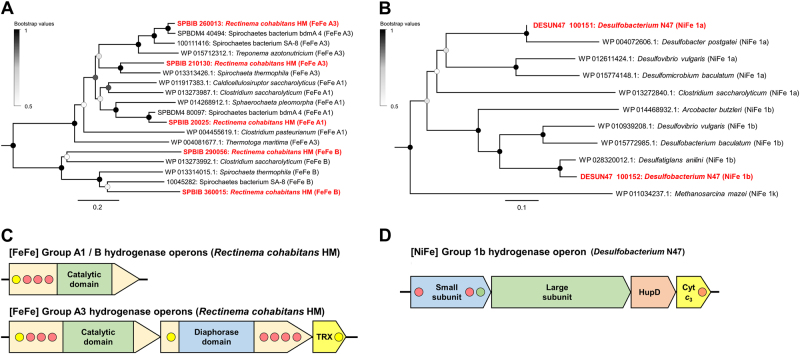
Fig. 2.
Determinants of H2 metabolism in Spirochaetes in hydrocarbon- and organohalide-contaminated environments. a Phylogenetic tree of the [FeFe]-hydrogenase catalytic subunit sequences detected in R. cohabitans, uncultured Spirochete bacterium bdmA 4, and uncultured Spirochete bacterium SA-8. b Phylogenetic tree of the [NiFe]-hydrogenase catalytic subunit sequences detected in Desulfobacterium N47. c Genetic organization of hydrogenases from R. cohabitans. d Genetic organization of the hydrogenase operon detected in Desulfobacterium N47. The illustrated tree for Figure 2a is condensed, with the full tree including all hydrogenases in Figure S1. The genetic organization of the Group 1a [NiFe] hydrogenase gene is not shown due to incomplete sequence coverage. Genetic organization diagrams are shown to scale and genes/domains are color-coded as follows: green, catalytic site; blue, secondary subunit; yellow, electron acceptor or donor; light orange, maturation factor. Redox-active centers are shown in circles, where yellow indicates [2Fe2S] cluster, green [3Fe4S] cluster, and red [4Fe4S] cluster