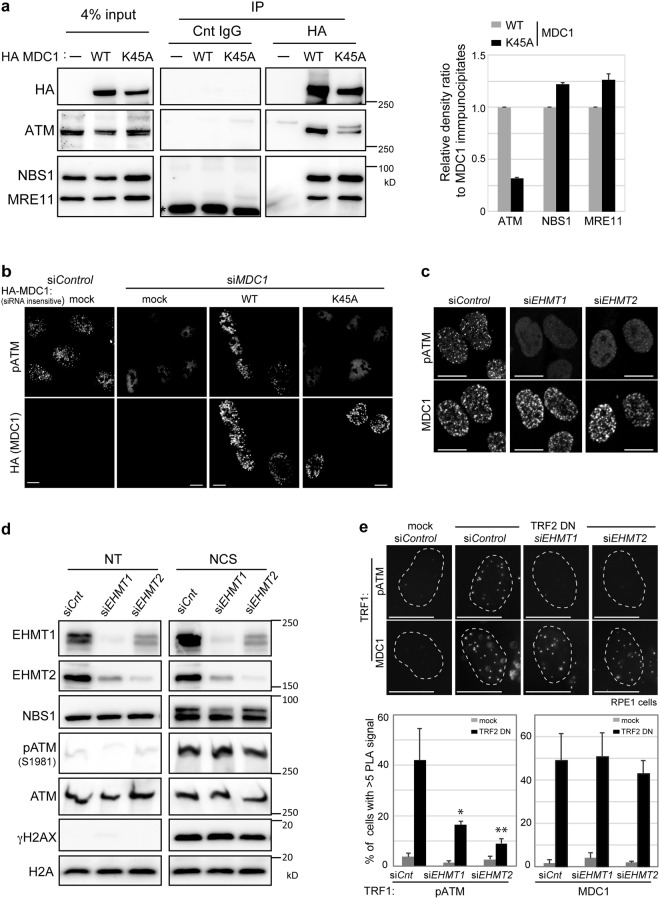
Figure 4.
MDC1 methylation is required for the amplification of activated ATM around the DNA double strand break sites. (a) Coimmunoprecipitation of MDC1 with ATM, NBS1 or MRE11 from U2OS cells transfected with HA-tagged MDC1 (WT or K45A). Asterisk: non-specific bands. Right panels: Quantification of immunoprecipitated ATM, NBS1 and MRE11 calculated from the signal intensities normalized against level of MDC1 immunoprecipitates. Error bars, SD from triplicate measurements. (b,c) Immunofluorescence analysis of U2OS cells transfected with indicated siRNA oligonucleotides, (b) and siRNA-resistant HA-tagged MDC1 (WT or K45A), using indicated antibodies at 2 h after exposure to NCS (50 ng/ml for 15 min). Scale bar, 10 μm. (d) Immunoblotting analysis of whole-cell extracts from U2OS cells transfected with control, EHMT1- or EHMT2-targeting siRNAs, using the indicated antibodies at 2 h after exposure to NCS (100 ng/ml for 30 min). (e) In situ PLA of RPE1 cells introduced with mock or TRF2-DN combined with or without the knockdown of EHMT1 or EHMT2, subjected to immunoreaction using TRF1 antibodies combined with MDC1 or phosphor-ser1981 ATM antibodies. Representative images are shown in the left panel. Localization of MDC1 or activated ATM at dysfunctional telomeres are quantified as the percentage of cells with more than 5 PLA signals in nuclei, each based on at least 150 cells from three independent experiments (right). Error bars represent SD. Statistical significance was calculated using two-tailed, unpaired t-test compared with control cells introduced with TRF2-DN; *P < 0.0005, **P < 0.0001. Scale bar, 10 μm.