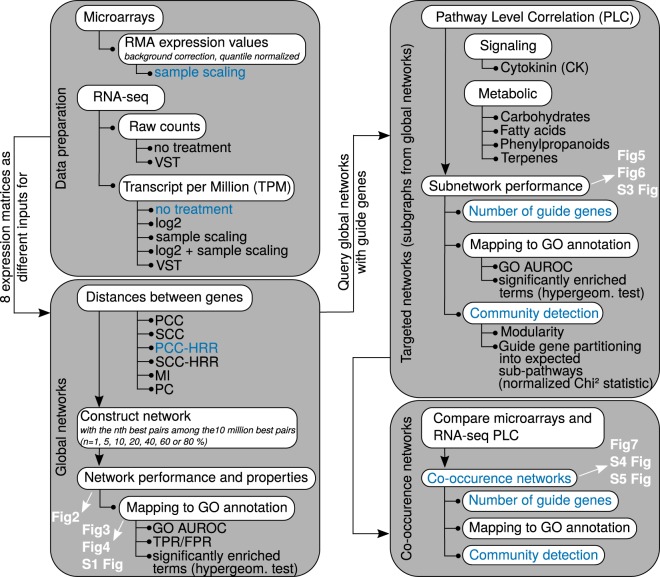
Figure 1.
Workflow for global and targeted network analyses. One microarray dataset and a RNA-seq dataset prepared according to 7 normalization procedures were used to generate eight expression matrices analyzed with six different distance measurements (Pearson’s or Spearman’s Correlation Coefficient, unranked or ranked with HRR, Mutual Information (MI) or Partial Correlations (PC)) to obtain 48 distance matrices. Each of these matrices was thresholded to obtain global networks at different confidence thresholds. Global networks were evaluated and also queried with specific guide gene sets reflecting 5 different pathways in a process named Pathway Level Correlation (PLC). The resulting subnetworks were evaluated and used to construct co-ocurrence networks between microarray and RNA-seq datasets. In white are indicated the figures corresponding to the different steps analyzed. Dataset × distance combinations are indicated in blue and characteristics that are improved by these combinations.