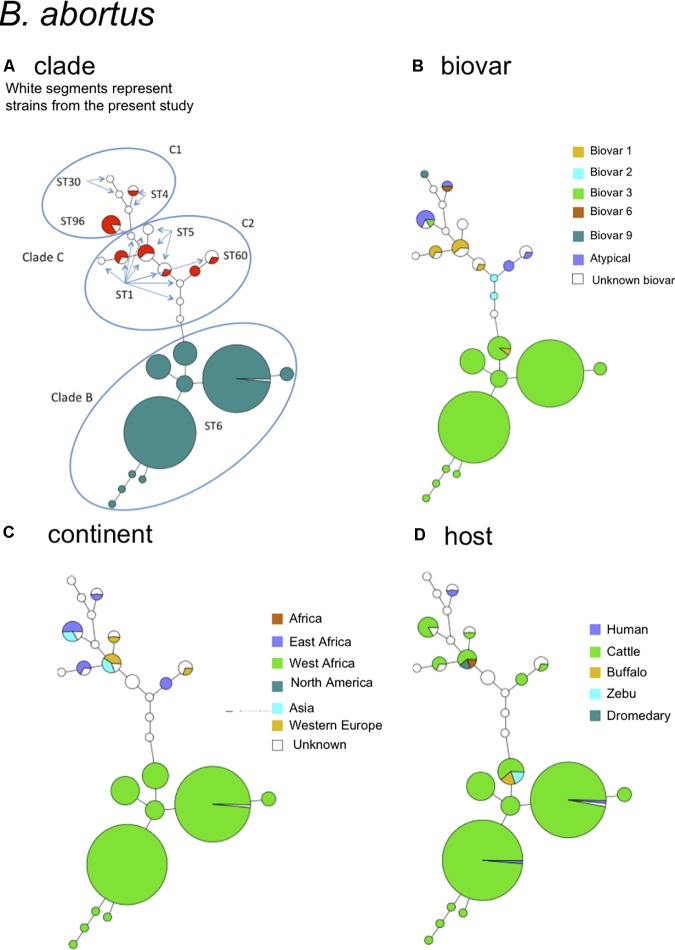
FIGURE 3.
B. abortus intraspecies diversity view provided by MLVA11. 232 entries are used including 213 B. abortus strains from this study and 19 representative entries from in silico data. The same MLVA11 minimum spanning tree was color-coded according to different characteristics. (A) Color coding according to assignment to B. abortus clade C or B. abortus clade B. Uncolored (white) entries correspond to in silico data. The MLST21 ST genotype for the white entries is indicated and was used to deduce the A, B, or C clade assignment defined by Whatmore et al. (2016). A tentative assignment of clades C1 and C2 is also proposed. (B) Color coding according to biovar (white when unknown). (C) Color-coding according to geographic origin defined at continent level (white when unknown). (D) Color-coding according to host (white when unknown).