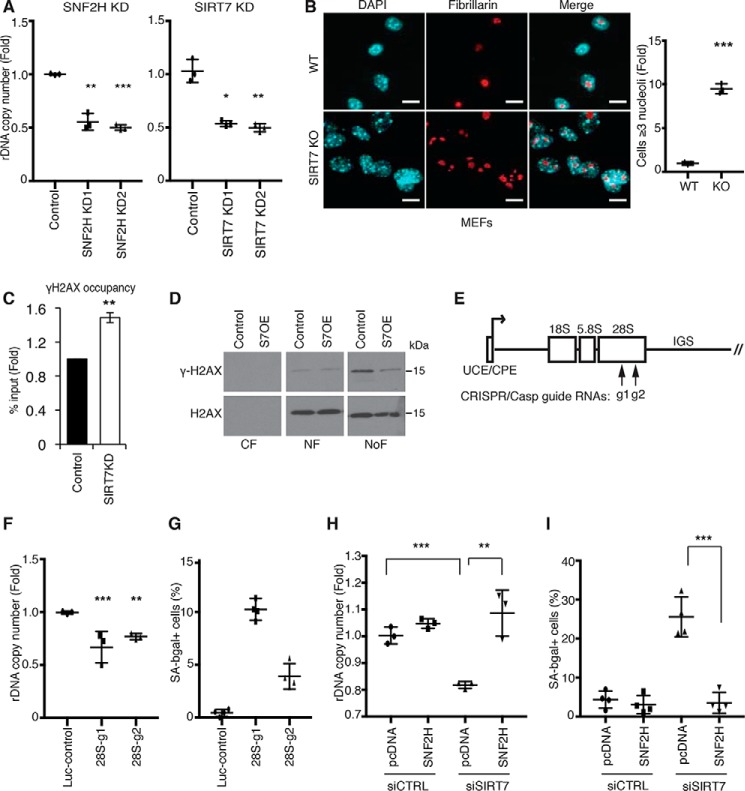
Figure 2.
rDNA instability is a driver of mammalian cellular senescence and is regulated by SIRT7. A, decreased rDNA copy number in SIRT7 KD or SNF2H KD cells. Data show two biological replicates. B, representative confocal microscopy images showing nucleolar fragmentation in SIRT7 KO MEFs. Nucleoli are stained with anti-fibrillarin; nuclear DNA is stained with DAPI. Scale bars, 15 μm. C, ChIP-qPCR showing increased γ-H2AX levels at rDNA promoters in SIRT7 KD cells. Data show the average of three biological replicates. D, immunoblots showing γ-H2AX levels in cytoplasmic (CF), nuclear (NF), and nucleolar (NoF) biochemical fractions of U2OS cells overexpressing SIRT7 (S7OE) or vector control. E, schematic of a human rDNA repeat. Arrows indicate target locations of the g1 and g2 CRISPR/Cas9 guide RNAs. F and G, decreased rDNA copy number (F) and increased senescence (G) in clonal cell lines following rDNA break induction with g1, g2, or luciferase control guide RNAs. Error bars indicate S.D. *, p < 0.05; **, p < 0.01 (one-tailed Student's t test). H and I, reversal of rDNA copy number loss (H) and cellular senescence (I) by overexpression of SNF2H in cells depleted of SIRT7 by siRNA transfection. Error bars indicate S.D. **, p < 0.01; ***, p < 0.001(one-tailed Student's t test). In all other panels, error bars indicate S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-tailed Student's t test). Graphs are representative of two (A, F, and H) or three (B) biologic replicates.