Figure 6.
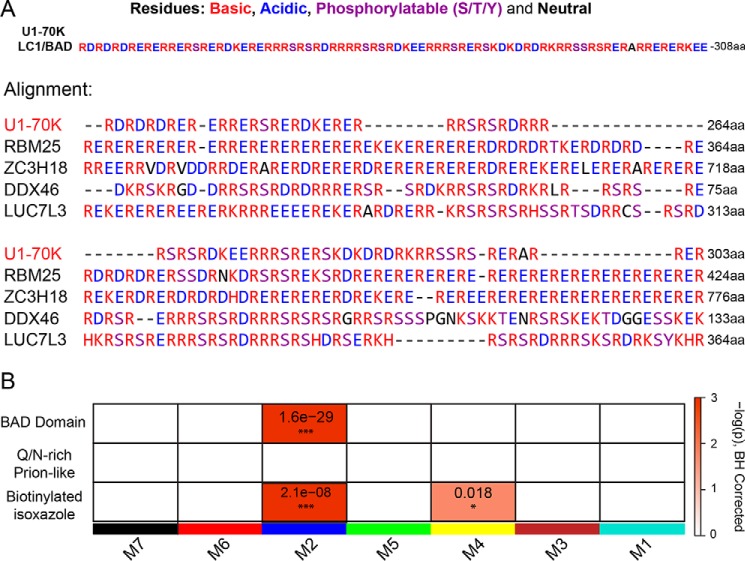
mRNA-processing module is enriched with structurally similar RNA-binding proteins containing BAD domains. A, LC1/BAD domain of U1-70K (residues 231–308) contains highly repetitive dipeptide repeats of basic (Arg/Lys) and acidic (Asp/Glu) residues. A list of 255 proteins that shared greater than 20% similarity to the LC1/BAD domain of U1-70K (E-values less than 0.005) was created using the Uniprot protein BLAST feature. Using Clustal Omega, an alignment was performed on the U1-70K LC1/BAD domain and the four most structurally similar proteins to highlight the repetitive basic and acidic residues in the sequence. Residues that can be phosphorylated (Ser/Thr/Tyr) are also highlighted as they can be negatively charged after modification. B, one-tailed Fisher's exact test was used to assess structural overlap of LC1-like BAD proteins from BLAST analysis with module membership for U1-70K-interacting partners (upper panel). The same analysis was repeated using Gln/Asn-rich prion-like RNA-binding proteins (middle panel) or RNA-binding proteins that were precipitated from nuclear extracts using biotin–isoxazole compound (bottom panel). Benjamini-Hochberg multiple comparison corrected p values for the module enrichment are highlighted. Significance is demonstrated by the color scales, which go from 0 (white) to ≥3 (red), representing −log(p).