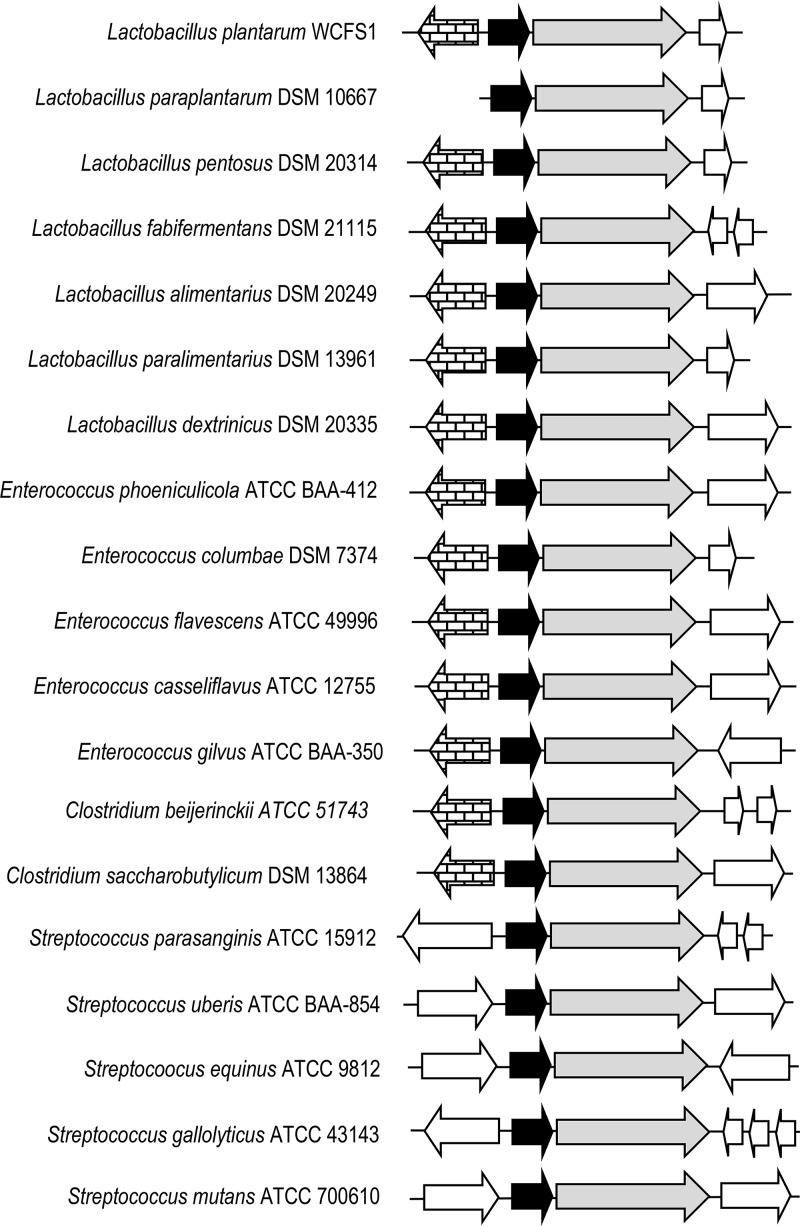
FIG 8.
Genetic organization of hydroxycinnamate reductases from several lactic acid bacteria, such as Lactobacillus plantarum WCFS1 (NC_004567.2), Lactobacillus paraplantarum DSM 10667 (NZ_CP013130.1), Lactobacillus pentosus DSM 20314 (GCA_001433755.1), Lactobacillus fabifermentans DSM 21115 (NZ_AYGX00000000.2), Lactobacillus alimentarius DSM 20249 (NZ_AZDQ00000000.1), Lactobacillus paralimentarius DSM 13961 (NZ_AZDH00000000.1), Lactobacillus dextrinicus DSM 20335 (NZ_AYYK00000000.1), Enterococcus phoeniculicola ATCC BAA-412 (NZ_ASWE00000000.1), Enterococcus columbae DSM 7374 (NZ_AHYW00000000.1), Enterococcus flavescens ATCC 49996 (NC_020995.1), Enterococcus casseliflavus ATCC 12755 (GCA_000191365.1), Enterococcus gilvus ATCC BAA-350 (NZ_ASWH00000000.1), Streptococcus parasanguinis ATCC 15912 (NC_015678.1), Streptococcus uberis ATCC BAA-854 (NC_012004.1), Streptococcus equinus ATCC 9812 (NZ_JNLO00000000.1), Streptococcus gallolyticus ATCC 43143 (NZ_CP018822.1), Streptococcus mutans ATCC 700610 (NC_004350.2), Clostridium beijerinckii ATCC 51743 (NZ_CP010086.2), and Clostridium saccharobutylicum DSM 13864 (NC_022571.1). Arrows indicate genes. Genes having putative identical functions are represented by identical shading. The genes having brick-like shading encode putative transcriptional regulators. Genes coding for putative reductases similar to HcrA and HcrB are represented by black and gray arrows, respectively.