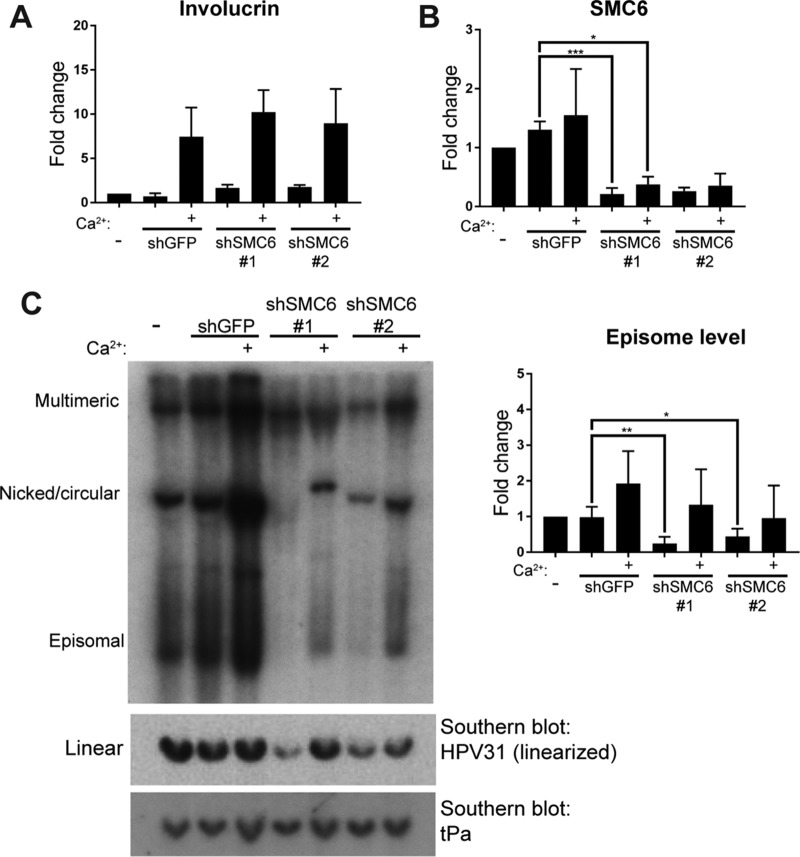
FIG 7.
SMC6 is not required for differentiation-dependent amplification of viral DNA. CIN612-9E cells were lentivirally transduced with control or SMC6 shRNA on day 0. Uninfected cells were collected at day 0. At 3 days postinfection, undifferentiated cells were harvested for analysis by qRT-PCR and Southern blotting or underwent 96 h of differentiation in high-calcium medium. (A) qRT-PCR measured involucrin levels as a marker of keratinocyte differentiation. (B) SMC6 mRNA levels were measured by qRT-PCR in CIN612-9E cells lentivirally transduced with SMC6 shRNA or GFP shRNA. All qRT-PCR data are normalized to those for cyclophilin A (n = 4). (C) (Left) Southern blotting detected HPV31 DNA from CIN612-9E cells. (Top) Total DNA was digested with BamHI targeting only mammalian DNA for visualization of the various forms of viral DNA. (Middle) Alternatively, DNA was digested with XbaI to linearize viral DNA (8 kbp). (Bottom) The Southern blot containing XbaI-digested DNA was probed for the human tPa gene as a loading control (1 to 2 kbp). (Right) Densitometric analysis of the episomal form of DNA as visualized by Southern blotting. P values were calculated using Student's t test. ***, P < 0.0001; **, P < 0.001; *, P < 0.05.