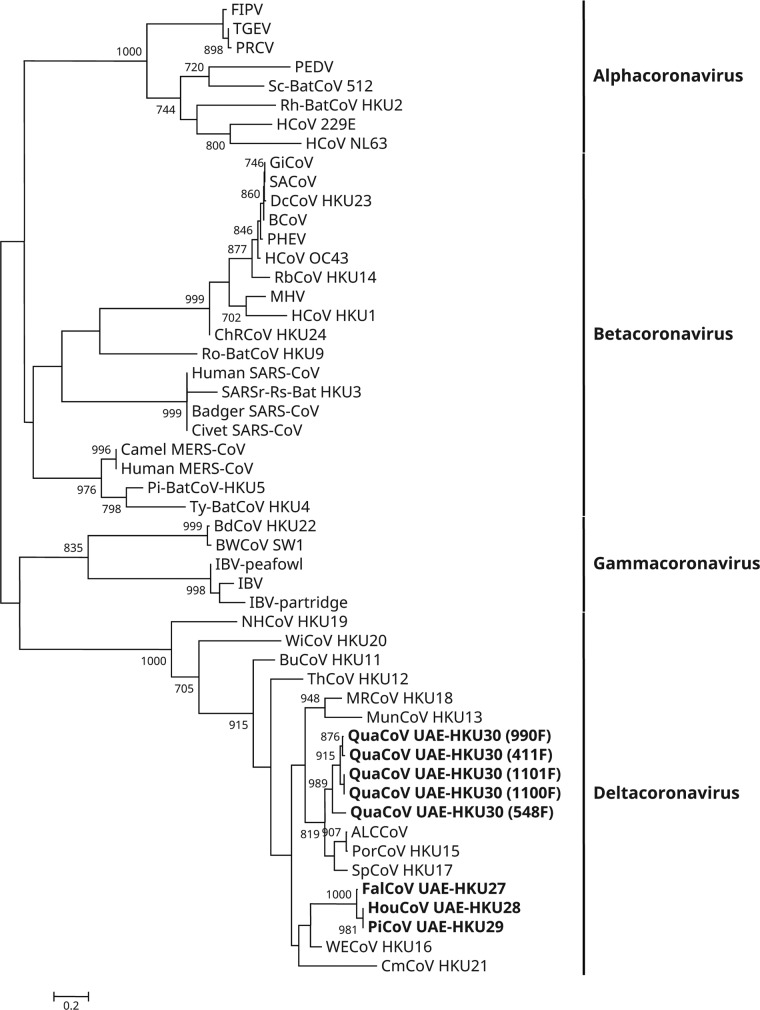
FIG 1.
Phylogenetic analysis of amino acid sequences of the 371-bp fragments (excluding primer sequences) of RNA-dependent RNA polymerases (RdRps) of coronaviruses (CoVs) identified from birds from Dubai in the present study. The tree was reconstructed by the maximum-likelihood method using PhyML 3.0 with the substitution model general time reversible with gamma distributed rate variation and estimated proportion of invariable sites (GTR+G+I). Bootstrap values were calculated from 1,000 trees. The scale bar indicates the number of nucleotide substitutions per site. The eight newly identified coronaviruses are shown in bold. The viruses and their respective DDBJ/ENA/GenBank accession numbers are as follow: ALCCoV, Asian leopard cat coronavirus (EF584908); Badger SARS-CoV, SARS-related Chinese ferret badger CoV (AY545919); BCoV, bovine CoV (NC_003045); BdCoV HKU22, bottlenose dolphin CoV HKU22 (KF793826); BuCoV HKU11, bulbul CoV HKU11 (FJ376619); BWCoV SW1, Beluga whale CoV SW1 (NC_010646); Camel MERS-CoV, camel Middle East respiratory syndrome CoV (KT751244); ChRCoV HKU24, China Rattus CoV HKU24 (KM349742); Civet SARS-CoV, SARS-related palm civet CoV (AY304488); CMCoV HKU21, common-moorhen CoV HKU21 (NC_016996); DcCoV HKU23, dromedary camel CoV HKU23 (KF906251); FalCoV UAE-HKU27, falcon CoV UAE-HKU27; FIPV, feline infectious peritonitis virus (AY994055); GiCoV, giraffe CoV (EF424622); HouCoV UAE-HKU28, houbara CoV UAE-HKU28; HCoV 229E, human CoV 229E (NC_002645); HCoV HKU1, human CoV HKU1 (NC_006577); HCoV NL63, human CoV NL63 (NC_005831); HCoV OC43, human CoV OC43 (NC_005147); Human MERS-CoV, human Middle East respiratory syndrome CoV (JX869059); Human SARS-CoV, severe acute respiratory syndrome-related human CoV (NC_004718); IBV, infectious bronchitis virus (NC_001451); IBV-partridge, partridge coronavirus (AY646283); IBV-peafowl, peafowl coronavirus (AY641576); MHV, murine hepatitis virus (NC_001846); MRCoV HKU18, magpie robin CoV HKU18 (NC_016993); MunCoV HKU13, munia CoV HKU13 (FJ376622); NHCoV HKU19, night-heron CoV HKU19 (NC_016994); PEDV, porcine epidemic diarrhea virus (NC_003436); PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); PiCoV UAE-HKU29, pigeon CoV UAE-HKU29; Pi-BatCoV HKU5, Pipistrellus bat CoV HKU5 (NC_009020); PorCoV HKU15, porcine CoV HKU15 (NC_016990); PRCV, porcine respiratory CoV (DQ811787); QuaCoV UAE-HKU30, quail CoV UAE-HKU30; RbCoV HKU14, rabbit CoV HKU14 (JN874559); Rh-BatCoV HKU2, Rhinolophus bat CoV HKU2 (EF203064); Ro-BatCoV HKU9, Rousettus bat CoV HKU9 (NC_009021); SACoV, sable antelope CoV (EF424621); SARSr-Rs-BatCoV HKU3, SARS-related Rhinolophus bat CoV HKU3 (DQ022305); Sc-BatCoV 512, Scotophilus bat CoV 512 (NC_009657); SpCoV HKU17, sparrow CoV HKU17 (NC_016992); TGEV, transmissible gastroenteritis virus (AJ271965); ThCoV HKU12, thrush CoV HKU12 (FJ376621); Ty-BatCoV HKU4, Tylonycteris bat CoV HKU4 (NC_009019); WECoV HKU16, white-eye CoV HKU16 (NC_016991); WiCoV HKU20, wigeon CoV HKU20 (NC_016995).