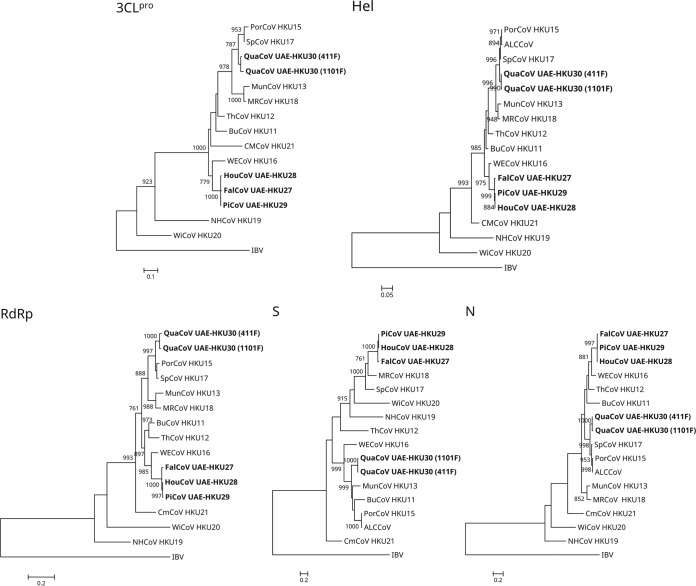
FIG 3.
Phylogenetic analyses of chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), helicase (Hel), spike (S) protein, and nucleocapsid (N) protein of falcon CoV-HKU27, houbara CoV-HKU28, pigeon CoV-HKU29, and quail CoV-HKU30. The trees were reconstructed by the maximum-likelihood method using PhyML 3.0 with the following substitution models: Le and Gascuel (LG) with gamma distributed rate variation (G) (3CLpro); LG with G, estimated proportion of invariable sites (I), and empirical frequencies (F) (RdRp); LG+G+F (Hel and N); and Whelan and Goldman (WAG)+G+I+F (S). Bootstrap values were calculated from 1,000 trees, and 314, 944, 599, 1,561, and 392 amino acid positions in 3CLpro, RdRp, Hel, S, and N, respectively, were included in the analyses. The scale bars indicate the number of amino acid substitutions per site. Viruses characterized in this study are in bold. Abbreviations for the viruses are the same as those in Fig. 1.